3RLC
 
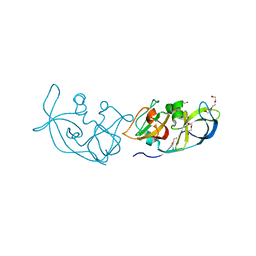 | |
3R6Y
 
 | | Crystal structure of chymotrypsin-treated aspartase from Bacillus sp. YM55-1 | | Descriptor: | Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
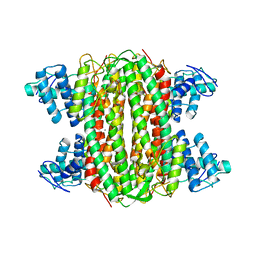
3TFI
 
 | | DMSP-dependent demethylase from P. ubique - with substrate DMSP | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, GLYCEROL, GcvT-like Aminomethyltransferase protein, ... | | Authors: | Schuller, D.J, Reisch, C.R, Moran, M.A, Whitman, W.B, Lanzilotta, W.N. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique.
Protein Sci., 21, 2012
|
|
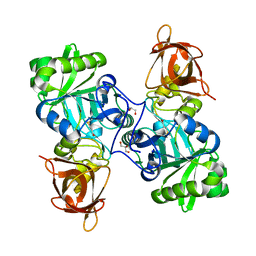
5M42
 
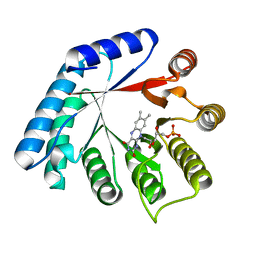 | | Structure of Thermus thermophilus L-proline dehydrogenase lacking alpha helices A, B and C | | Descriptor: | FLAVIN MONONUCLEOTIDE, Proline dehydrogenase | | Authors: | Martinez-Julvez, M, Huijbers, M.M.E, van Berkel, W.J.H, Medina, M. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Proline dehydrogenase from Thermus thermophilus does not discriminate between FAD and FMN as cofactor.
Sci Rep, 7, 2017
|
|
1WZA
 
 | |
1XE4
 
 | | Crystal Structure of Weissella viridescens FemX (K36M) Mutant | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Arthur, M, Mayer, C. | | Deposit date: | 2004-09-09 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of the UDP-MurNAc-Pentapeptide-Binding Cavity of the FemX Alanyl Transferase from Weissella viridescens
J.Bacteriol., 187, 2005
|
|
5NH6
 
 | |
5NH5
 
 | |
5NH8
 
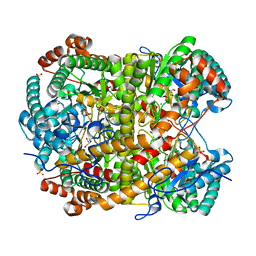 | |
5NHM
 
 | |
5NHE
 
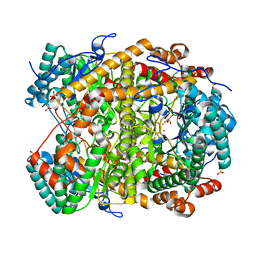 | |
1XC1
 
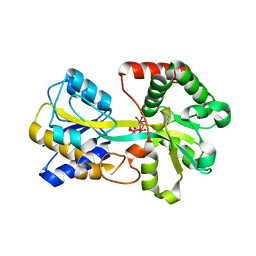 | | Oxo Zirconium(IV) Cluster in the Ferric Binding Protein (FBP) | | Descriptor: | OXO ZIRCONIUM(IV) CLUSTER, periplasmic iron-binding protein | | Authors: | Zhong, W, Alexeev, D, Harvey, I, Guo, M, Hunter, D.J.B, Zhu, H, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2004-08-31 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Assembly of an Oxo-Zirconium(IV) Cluster in a Protein Cleft
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1XFH
 
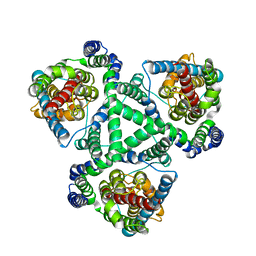 | |
1XF8
 
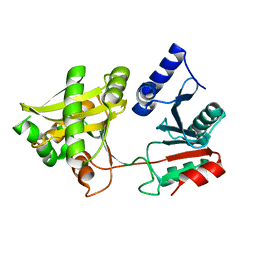 | | Crystal Structure of Weissella viridescens FemX (Y254F) Mutant | | Descriptor: | FemX, MAGNESIUM ION | | Authors: | Biarrotte-Sorin, S, Maillard, A.P, Arthur, M, Mayer, C. | | Deposit date: | 2004-09-14 | | Release date: | 2005-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Site-Directed Mutagenesis of the UDP-MurNAc-Pentapeptide-Binding Cavity of the FemX Alanyl Transferase from Weissella viridescens
J.BACTERIOL., 187, 2005
|
|
3QQP
 
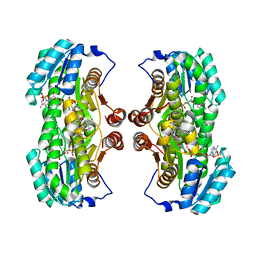 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1XIX
 
 | |
5NHB
 
 | |
5NHA
 
 | |
5N7J
 
 | |
5NH9
 
 | |
5N6V
 
 | | Crystal structure of Neisseria polysaccharea amylosucrase mutant derived from Neutral genetic Drift-based engineering | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amylosucrase, ... | | Authors: | Daude, D, Verges, A, Tranier, S. | | Deposit date: | 2017-02-16 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutral Genetic Drift-Based Engineering of a Sucrose-Utilizing Enzyme toward Glycodiversification.
Acs Catalysis, 2019
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
3RLK
 
 | |
5NH7
 
 | |
5NH4
 
 | |
