9BF6
 
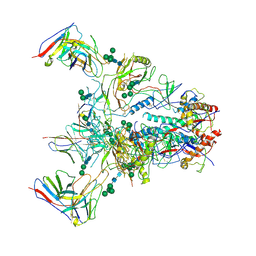 | |
9BEW
 
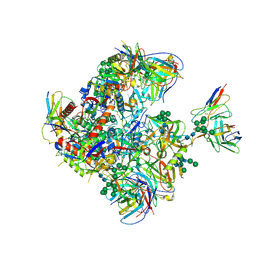 | |
9BER
 
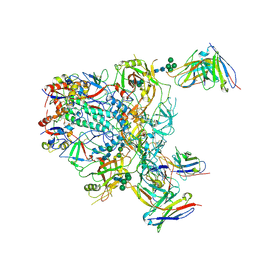 | | Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Design of soluble HIV-1 envelope trimers free of covalent gp120-gp41 bonds with prevalent native-like conformation.
Cell Rep, 43, 2024
|
|
9BF0
 
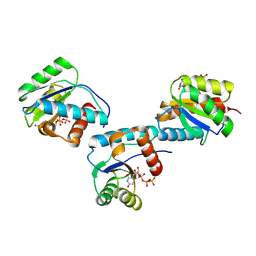 | | MID domain of human Argo2 bound to UTP | | Descriptor: | Protein argonaute-2, URIDINE 5'-TRIPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BF2
 
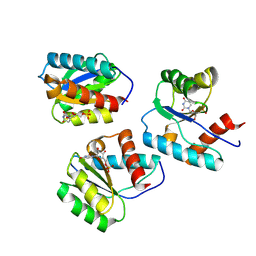 | | MID domain of Ago2 bound to UMP | | Descriptor: | Protein argonaute-2, URIDINE-5'-MONOPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BEZ
 
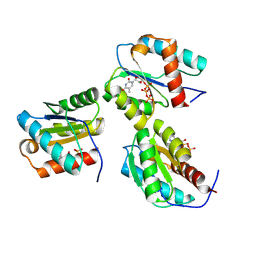 | | MID domain of human Argo2 bound to RNA | | Descriptor: | Protein argonaute-2, [(3~{S},4~{R},5~{R})-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-4-oxidanyl-oxolan-3-yl] [oxidanyl(phosphonooxy)phosphoryl] hydrogen phosphate | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BE2
 
 | |
9BDN
 
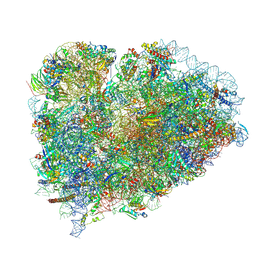 | |
9BDP
 
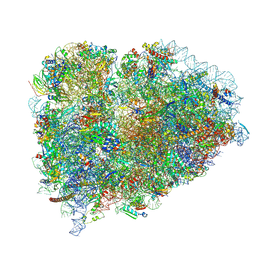 | |
9BDL
 
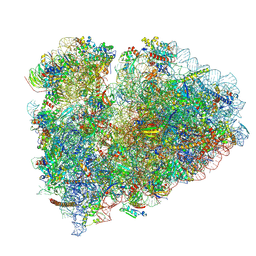 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
9EZJ
 
 | | Apo human TDO in complex with a bound inhibitor (Cpd-4) | | Descriptor: | Tryptophan 2,3-dioxygenase, alpha-methyl-L-tryptophan, ethyl (9~{R})-2-methoxy-4-oxidanylidene-9-[[(1~{S})-1-phenylethyl]-[(2-propan-2-ylphenyl)carbamoyl]amino]-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidine-3-carboxylate | | Authors: | Wicki, M, Mac Sweeney, A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Discovery and binding mode of small molecule inhibitors of the apo form of human TDO2
To Be Published
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDU
 
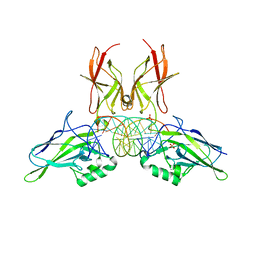 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
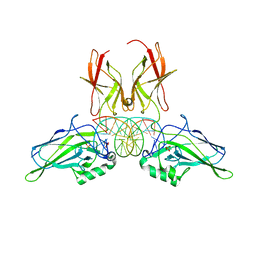 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDW
 
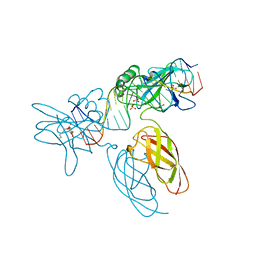 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDD
 
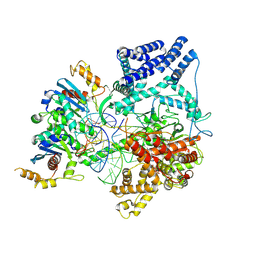 | | Cryo-EM Structure of Non-Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
8Z1L
 
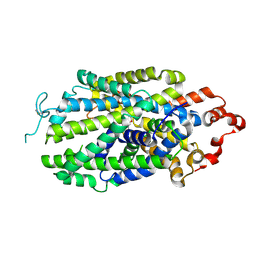 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
9BDC
 
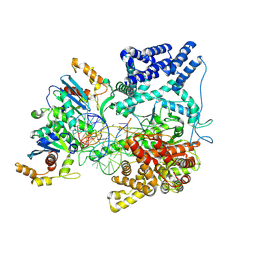 | | Cryo-EM Structure of the TEFM bound Human Mitochondrial Transcription Elongation Complex in a Closed Fingers Domain Conformation | | Descriptor: | DNA-directed RNA polymerase, mitochondrial, Non-Template Strand DNA (NT27mt_+1T), ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
9EZ2
 
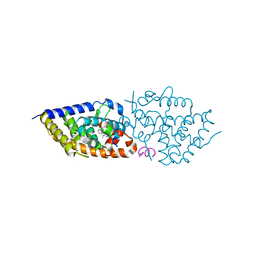 | | Vitamin D receptor complex with 1,4b,25-trihydroxyvitamin D3 | | Descriptor: | 1,4b,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
9BD3
 
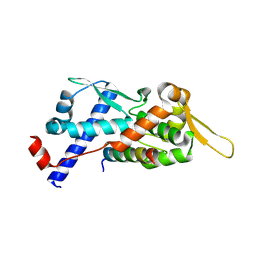 | | Structure of the MAGEA4 MHD-RAD18 R6BD Complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Melanoma antigen A 4 | | Authors: | Forker, K, Zhou, P. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of MAGEA4 MHD-RAD18 R6BD reveals a flipped binding mode compared to AlphaFold2 prediction.
Embo J., 43, 2024
|
|
9EZ1
 
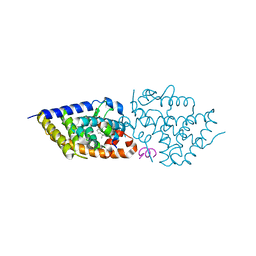 | | Vitamin D receptor in complex with 1,4a,25-trihydroxyvitamin D3 | | Descriptor: | 1,4a,25-trihydroxyvitamin D3, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2024-04-10 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-Hydroxy-1 alpha ,25-Dihydroxyvitamin D 3 : Synthesis and Structure-Function Study.
Biomolecules, 14, 2024
|
|
9BCT
 
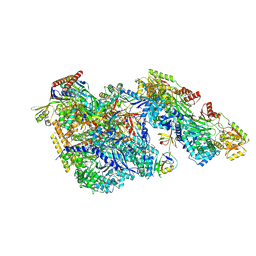 | |
9EXY
 
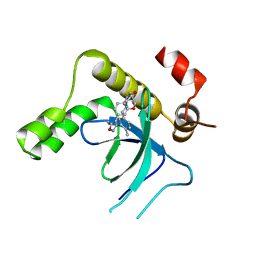 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 34. | | Descriptor: | 7-[5-methyl-3-[2-methyl-5-(piperidin-1-ylmethyl)phenyl]-1,2-oxazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
9BCU
 
 | |
9EXW
 
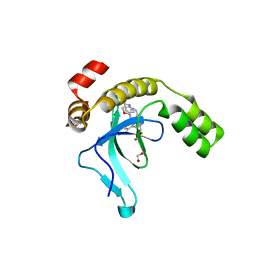 | | Crystal structure of the PWWP1 domain of NSD2 bound by compound 17. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-methyl-5-[2-methyl-5-[(pyridin-3-ylamino)methyl]phenyl]imidazol-4-yl]-4~{H}-1,4-benzoxazin-3-one, Histone-lysine N-methyltransferase NSD2 | | Authors: | Collie, G.W. | | Deposit date: | 2024-04-09 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Novel Potent NSD2-PWWP1 Ligands Using Structure-Based Design and Computational Approaches.
J.Med.Chem., 67, 2024
|
|
