5JFB
 
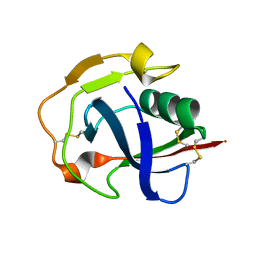 | | Crystal structure of the scavenger receptor cysteine-rich domain 5 (SRCR5) from porcine CD163 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Jiang, L, Qiao, S, Zhang, G, Li, R. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Fifth Scavenger Receptor Cysteine-Rich Domain of Porcine CD163 Reveals an Important Residue Involved in Porcine Reproductive and Respiratory Syndrome Virus Infection
J. Virol., 91, 2017
|
|
7DPX
 
 | |
8H7J
 
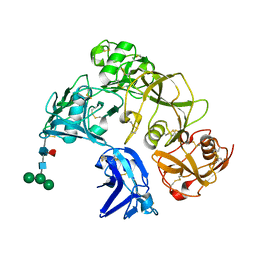 | | The crystal structure of CD163 SRCR5-9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor cysteine-rich type 1 protein M130, ... | | Authors: | Luo, Z.P, Li, R, Ma, H.F. | | Deposit date: | 2022-10-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of CD163 SRCR5-9
To Be Published
|
|
1BY2
 
 | |
5HRJ
 
 | |
5A2E
 
 | | Extracellular SRCR domains of human CD6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-CELL DIFFERENTIATION ANTIGEN CD6 | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
8J8D
 
 | |
8J8T
 
 | |
7C00
 
 | | Crystal structure of the SRCR domain of human SCARA5. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
7BZZ
 
 | | Crystal structure of the SRCR domain of mouse SCARA5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Scavenger receptor class A member 5 | | Authors: | Yu, B, He, Y. | | Deposit date: | 2020-04-29 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Interactions of ferritin with scavenger receptor class A members.
J.Biol.Chem., 295, 2020
|
|
2OYA
 
 | | Crystal structure analysis of the dimeric form of the SRCR domain of mouse MARCO | | Descriptor: | Macrophage receptor MARCO, SULFATE ION | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
2OY3
 
 | | Crystal structure analysis of the monomeric SRCR domain of mouse MARCO | | Descriptor: | MAGNESIUM ION, Macrophage receptor MARCO | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
2JP0
 
 | | Solution structure of the N-terminal extraceullular domain of the lymphocyte receptor CD5 calculated using inferential structure determination (ISD) | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Garza-Garcia, A, Harris, R, Esposito, D, Driscoll, P.C, Rieping, W. | | Deposit date: | 2007-04-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and conformational plasticity of the N-terminal scavenger receptor cysteine-rich domain of human CD5
J.Mol.Biol., 378, 2008
|
|
2JOP
 
 | |
6SAN
 
 | | SALSA / DMBT1 / GP340 SRCR domain 8 soaked in calcium and magnesium | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Johnson, S, Loimaranta, V, Lea, S.M. | | Deposit date: | 2019-07-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6SA5
 
 | | SALSA / DMBT1 / GP340 SRCR domain 8 | | Descriptor: | Deleted in malignant brain tumors 1 protein, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Reichhardt, M.P, Johnson, S, Loimaranta, V, Lea, S.M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6SA4
 
 | | SALSA / DMBT1 / GP340 SRCR domain 1 | | Descriptor: | CHLORIDE ION, Deleted in malignant brain tumors 1 protein, GLYCEROL, ... | | Authors: | Reichhardt, M.P, Lea, S.M, Johnson, S. | | Deposit date: | 2019-07-16 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structures of SALSA/DMBT1 SRCR domains reveal the conserved ligand-binding mechanism of the ancient SRCR fold.
Life Sci Alliance, 3, 2020
|
|
6J02
 
 | | Crystal structure of the SRCR domain of mouse SCARA1 | | Descriptor: | CALCIUM ION, Macrophage scavenger receptor types I and II | | Authors: | Cheng, C, Hu, Z. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | The scavenger receptor SCARA1 (CD204) recognizes dead cells through spectrin.
J.Biol.Chem., 294, 2019
|
|
6K0L
 
 | | The crystal structure of simian CD163 SRCR5 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
6H8M
 
 | |
6K0O
 
 | | The crystal structure of human CD163-like homolog SRCR8 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M160 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
5ZE3
 
 | |
8WYS
 
 | | Local map of human CD5L bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Y.X, Su, C, Xiao, J.Y. | | Deposit date: | 2023-10-31 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | CD5L associates with IgM via the J chain.
Nat Commun, 15, 2024
|
|
8WYR
 
 | | Cryo-EM structure of human CD5L bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Y.X, Su, C, Xiao, J.Y. | | Deposit date: | 2023-10-31 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | CD5L associates with IgM via the J chain.
Nat Commun, 15, 2024
|
|
2XRC
 
 | | Human complement factor I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HUMAN COMPLEMENT FACTOR I | | Authors: | Roversi, P, Johnson, S, Lea, S.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Basis for Complement Factor I Control and its Disease-Associated Sequence Polymorphisms.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
