3D0Z
 
 | |
3CRU
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
3CRT
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
4LKU
 
 | |
3INO
 
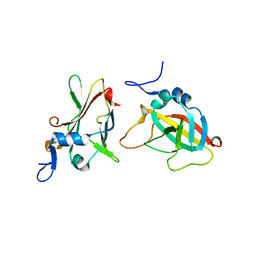 | | 1.95A Resolution Structure of Protective Antigen Domain 4 | | Descriptor: | Protective antigen PA-63 | | Authors: | Lovell, S, Williams, A.S, Anbanandam, A, El-Chami, R, Bann, J.G. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Domain 4 of the anthrax protective antigen maintains structure and binding to the host receptor CMG2 at low pH
Protein Sci., 18, 2009
|
|
4GJ4
 
 | |
4B09
 
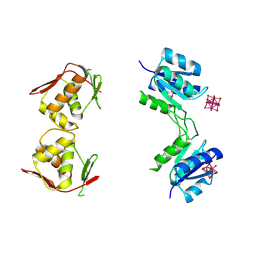 | | Structure of unphosphorylated BaeR dimer | | Descriptor: | HEXATANTALUM DODECABROMIDE, TRANSCRIPTIONAL REGULATORY PROTEIN BAER | | Authors: | Choudhury, H, Beis, K. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Dimeric Form of the Unphosphorylated Response Regulator Baer.
Protein Sci., 22, 2013
|
|
2RT6
 
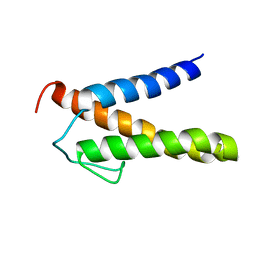 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for PriC N-terminal domain | | Descriptor: | Primosomal replication protein N'' | | Authors: | Aramaki, T, Abe, Y, Katayama, T, Ueda, T. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of a replication restart primosome factor, PriC, in Escherichia coli.
Protein Sci., 22, 2013
|
|
4M2T
 
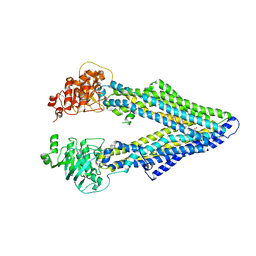 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-SSS | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
4MSP
 
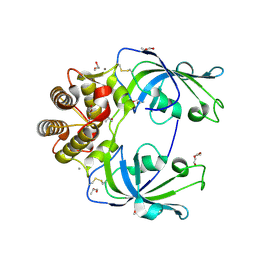 | | Crystal structure of human peptidyl-prolyl cis-trans isomerase FKBP22 (aka FKBP14) containing two EF-hand motifs | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase FKBP14, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2013-09-18 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human peptidyl-prolyl cis-trans isomerase FKBP22 containing two EF-hand motifs.
Protein Sci., 23, 2014
|
|
3A4C
 
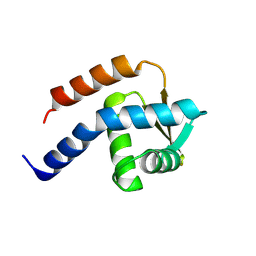 | | Crystal structure of cdt1 C terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Cho, Y, Lee, J.H. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
4M1M
 
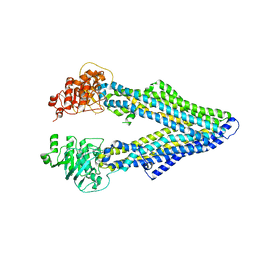 | |
4M2S
 
 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
4LQ8
 
 | |
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
2KLO
 
 | | Structure of the Cdt1 C-terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Khayrutdinov, B.I, Bae, W.J, Yun, Y.M, Tsuyama, T, Kim, J.J, Hwang, E, Ryu, K.-S, Cheong, H.-K, Cheong, C, Karplus, P.A, Guntert, P, Tada, S, Jeon, Y.H, Cho, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
2M8E
 
 | | NMR structure of the PAI subdomain of Sleeping Beauty transposase | | Descriptor: | SLEEPING BEAUTY TRANSPOSASE | | Authors: | Eubanks, C, Schreifels, J, Aronovich, E, Carlson, D, Hacjkett, P, Nesmelova, I. | | Deposit date: | 2013-05-17 | | Release date: | 2013-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of Sleeping Beauty transposase binding to DNA.
Protein Sci., 23, 2014
|
|
2L83
 
 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
4HRS
 
 | |
4J7W
 
 | | E3_5 DARPin L86A mutant | | Descriptor: | DARPin_E3_5_L86A | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4J8Y
 
 | | E3_5 DARPin D77S mutant | | Descriptor: | DARPin_E3_5_D77S | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-15 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4JP8
 
 | | Crystal structure of Pro-F17H/S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Yuzaki, K, You, D.J, Uehara, R, Koga, Y, Kanaya, S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Increase in activation rate of Pro-Tk-subtilisin by a single nonpolar-to-polar amino acid substitution at the hydrophobic core of the propeptide domain
Protein Sci., 22, 2013
|
|
4JE5
 
 | | Crystal structure of the aromatic aminotransferase Aro8, a putative alpha-aminoadipate aminotransferase in Saccharomyces cerevisiae | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic/aminoadipate aminotransferase 1, ... | | Authors: | Bulfer, S.L, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2013-02-26 | | Release date: | 2013-09-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae Aro8, a putative alpha-aminoadipate aminotransferase.
Protein Sci., 22, 2013
|
|
4KDT
 
 | | Structure of an early native-like intermediate of beta2-microglobulin amyloidosis | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Nanobody24, ... | | Authors: | Vanderhaegen, S, Fislage, M, Pardon, E, Versees, W, Steyaert, J. | | Deposit date: | 2013-04-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of an early native-like intermediate of beta 2-microglobulin amyloidogenesis.
Protein Sci., 22, 2013
|
|
4KYD
 
 | | Partial Structure of the C-terminal domain of the HPIV4B phosphoprotein, fused to MBP. | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Maltose-binding periplasmic protein, Phosphoprotein, ... | | Authors: | Yegambaram, K, Bulloch, E.M.M, Kingston, R.L. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Protein domain definition should allow for conditional disorder.
Protein Sci., 22, 2013
|
|
