1SJF
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Cobalt Hexammine solution | | Descriptor: | COBALT HEXAMMINE(III), Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A conformational switch controls hepatitis delta virus ribozyme catalysis.
Nature, 429, 2004
|
|
1SJG
 
 | | Solution Structure of T4moC, the Rieske Ferredoxin Component of the Toluene 4-Monooxygenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Toluene-4-monooxygenase system protein C | | Authors: | Skjeldal, L, Peterson, F.C, Doreleijers, J.F, Moe, L.A, Pikus, J.D, Volkman, B.F, Westler, W.M, Markley, J.L, Fox, B.G. | | Deposit date: | 2004-03-03 | | Release date: | 2004-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of T4moC, the Rieske ferredoxin component of the toluene 4-monooxygenase complex
J.Biol.Inorg.Chem., 9, 2004
|
|
1SJH
 
 | | HLA-DR1 complexed with a 13 residue HIV capsid peptide | | Descriptor: | Enterotoxin type C-3, GAG polyprotein, HLA class II histocompatibility antigen, ... | | Authors: | Zavala-Ruiz, Z, Strug, I, Walker, B.D, Norris, P.J, Stern, L.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SJI
 
 | | Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization | | Descriptor: | Calsequestrin, cardiac muscle isoform | | Authors: | Park, H.J, Park, I.Y, Kim, E.J, Youn, B, Fields, K, Dunker, A.K, Kang, C.H. | | Deposit date: | 2004-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization.
J.Biol.Chem., 279, 2004
|
|
1SJJ
 
 | |
1SJK
 
 | | A DUPLEX DNA WITH AN ABASIC SITE IN A DA TRACT, ALPHA FORM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*TP*ORPP*TP*TP*GP*CP*G)-3') | | Authors: | Wang, K.Y, Parker, S.A, Goljer, I, Bolton, P.H. | | Deposit date: | 1997-07-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a duplex DNA with an abasic site in a dA tract.
Biochemistry, 36, 1997
|
|
1SJL
 
 | | A DUPLEX DNA WITH AN ABASIC SITE IN A DA TRACT, BETA FORM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*TP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*AP*TP*TP*(AAB)P*TP*TP*GP*CP*G)-3') | | Authors: | Wang, K.Y, Parker, S.A, Goljer, I, Bolton, P.H. | | Deposit date: | 1997-07-22 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a duplex DNA with an abasic site in a dA tract.
Biochemistry, 36, 1997
|
|
1SJM
 
 | | Nitrite bound copper containing nitrite reductase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, COPPER (II) ION, ... | | Authors: | Tocheva, E.I, Rosell, F.I, Mauk, A.G, Murphy, M.E.P. | | Deposit date: | 2004-03-03 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side-on copper-nitrosyl coordination by nitrite reductase.
Science, 304, 2004
|
|
1SJN
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.S, Kim, M.-Y, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-03-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
1SJP
 
 | |
1SJQ
 
 | | NMR Structure of RRM1 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1SJR
 
 | | NMR Structure of RRM2 from Human Polypyrimidine Tract Binding Protein Isoform 1 (PTB1) | | Descriptor: | Polypyrimidine tract-binding protein 1 | | Authors: | Simpson, P.J, Monie, T.P, Szendroi, A, Davydova, N, Tyzack, J.K, Conte, M.R, Read, C.M, Cary, P.D, Svergun, D.I, Konarev, P.V, Petoukhov, M.V, Curry, S, Matthews, S.J. | | Deposit date: | 2004-03-04 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Interactions of the N-Terminal RRM Domains of PTB
Structure, 12, 2004
|
|
1SJS
 
 | |
1SJT
 
 | | MINI-PROINSULIN, TWO CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1SJU
 
 | | MINI-PROINSULIN, SINGLE CHAIN INSULIN ANALOG MUTANT: DES B30, HIS(B 10)ASP, PRO(B 28)ASP AND PEPTIDE BOND BETWEEN LYS B 29 AND GLY A 1, NMR, 20 STRUCTURES | | Descriptor: | PROINSULIN | | Authors: | Hua, Q.X, Hu, S.Q, Jia, W.H, Chu, Y.C, Burke, G.T, Wang, S.H, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 1997-10-09 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Mini-proinsulin and mini-IGF-I: homologous protein sequences encoding non-homologous structures.
J.Mol.Biol., 277, 1998
|
|
1SJV
 
 | | Three-Dimensional Structure of a Llama VHH Domain Swapping | | Descriptor: | r9 | | Authors: | Spinelli, S, Desmyter, A, Frenken, L, Verrips, T, Cambillau, C. | | Deposit date: | 2004-03-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure.
Febs Lett., 564, 2004
|
|
1SJW
 
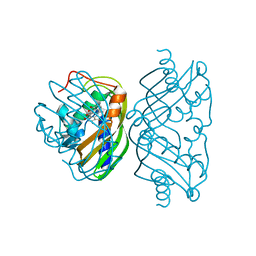 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
1SJX
 
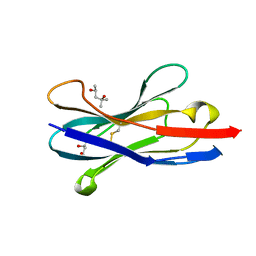 | | Three-Dimensional Structure of a Llama VHH Domain OE7 binding the cell wall protein Malf1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, immunoglobulin VH domain | | Authors: | Dolk, E, van der Vaart, M, Hulsik, D.L, Vriend, G, de Haard, H, Spinelli, S, Cambillau, C, Frenken, L, Verrips, T. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo.
Appl.Environ.Microbiol., 71, 2005
|
|
1SJY
 
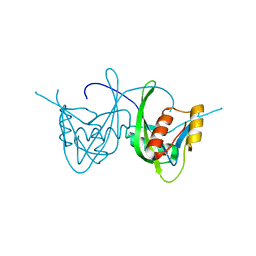 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SJZ
 
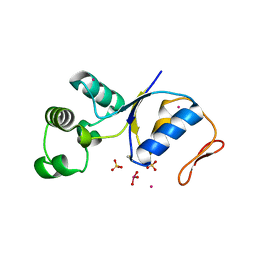 | | ARSENATE REDUCTASE R60K MUTANT +0.4M ARSENITE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION, ... | | Authors: | Demel, S, Edwards, B.F. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SK0
 
 | | ARSENATE REDUCTASE R60A MUTANT +0.4M ARSENITE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION, ... | | Authors: | Demel, S, Edwards, B.F. | | Deposit date: | 2004-03-04 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SK1
 
 | | ARSENATE REDUCTASE R60K MUTANT +0.4M ARSENATE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-03-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SK2
 
 | | ARSENATE REDUCTASE R60A MUTANT +0.4M ARSENATE FROM E. COLI | | Descriptor: | Arsenate reductase, CESIUM ION, SULFATE ION | | Authors: | DeMel, S, Edwards, B.F. | | Deposit date: | 2004-03-04 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product.
Protein Sci., 13, 2004
|
|
1SK3
 
 | | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | NICKEL (II) ION, Peptidoglycan recognition protein I-alpha, SULFATE ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
1SK4
 
 | | crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha | | Descriptor: | Peptidoglycan recognition protein I-alpha, SODIUM ION | | Authors: | Guan, R, Malchiodi, E.L, Qian, W, Schuck, P, Mariuzza, R.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Ialpha
J.Biol.Chem., 279, 2004
|
|
