1NSZ
 
 | |
1NT0
 
 | | Crystal structure of the CUB1-EGF-CUB2 region of MASP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Feinberg, H, Uitdehaag, J.C.M, Davies, J.M, Wallis, R, Drickamer, K, Weis, W.I. | | Deposit date: | 2003-01-28 | | Release date: | 2003-05-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2
Embo J., 22, 2003
|
|
1NT1
 
 | | thrombin in complex with selective macrocyclic inhibitor | | Descriptor: | (6R,21AS)-17-CHLORO-6-CYCLOHEXYL-2,3,6,7,10,11,19,20-OCTAHYDRO-1H,5H-PYRROLO[1,2-K][1,4,8,11,14]BENZOXATETRAAZA-CYCLOHEPTADECINE-5,8,12,21(9H,13H,21AH)-TETRONE, Hirudin, thrombin | | Authors: | Nantermet, P.G, Barrow, J.C, Newton, C.L, Pellicore, J.M, Young, M, Lewis, S.D, Lucas, B.J, Krueger, J.A, McMasters, D.R, Yan, Y, Kuo, L.C, Vacca, J.P, Selnick, H.G. | | Deposit date: | 2003-01-28 | | Release date: | 2003-09-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent and selective macrocyclic thrombin inhibitors
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NT2
 
 | | CRYSTAL STRUCTURE OF FIBRILLARIN/NOP5P COMPLEX | | Descriptor: | Fibrillarin-like pre-rRNA processing protein, S-ADENOSYLMETHIONINE, conserved hypothetical protein | | Authors: | Aittaleb, M, Rashid, R, Chen, Q, Palmer, J.R, Daniels, C.J, Li, H. | | Deposit date: | 2003-01-28 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and function of archaeal box C/D sRNP core proteins.
Nat.Struct.Biol., 10, 2003
|
|
1NT3
 
 | | HUMAN NEUROTROPHIN-3 | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Butte, M.J, Hwang, P.K, Mobley, W.C, Fletterick, R.J. | | Deposit date: | 1999-05-17 | | Release date: | 1999-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of neurotrophin-3 homodimer shows distinct regions are used to bind its receptors.
Biochemistry, 37, 1998
|
|
1NT4
 
 | | Crystal structure of Escherichia coli periplasmic glucose-1-phosphatase H18A mutant complexed with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-glucopyranose, Glucose-1-phosphatase | | Authors: | Lee, D.C, Cottrill, M.A, Forsberg, C.W, Jia, Z, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-01-28 | | Release date: | 2004-01-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase.
J.Biol.Chem., 278, 2003
|
|
1NT5
 
 | | F1-Gramicidin A in Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN A | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1NT6
 
 | | F1-Gramicidin C In Sodium Dodecyl Sulfate Micelles (NMR) | | Descriptor: | GRAMICIDIN C | | Authors: | Townsley, L.E, Fletcher, T.G, Hinton, J.F. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-11 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The Structure, Cation Binding, Transport, and Conductance of Gly15-Gramicidin a Incorporated Into Sds Micelles and Pc/Pg Vesicles.
Biochemistry, 42, 2003
|
|
1NT8
 
 | | Structural Characterisation of the Holliday junction formed by the sequence CCGGTACCGG at 2.00 A | | Descriptor: | 5'-d(CpCpGpGpTpApCpCpGpG)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-29 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of two Holliday Junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NT9
 
 | | Complete 12-subunit RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II LARGEST SUBUNIT, ... | | Authors: | Armache, K.-J, Kettenberger, H, Cramer, P. | | Deposit date: | 2003-01-29 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Architecture of initiation-competent 12-subunit RNA polymerase II
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NTA
 
 | | 2.9 A crystal structure of Streptomycin RNA-aptamer | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', BARIUM ION, ... | | Authors: | Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
1NTB
 
 | | 2.9 A crystal structure of Streptomycin RNA-aptamer complex | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
1NTC
 
 | |
1NTD
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE MUTANT M150E THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
1NTE
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE SECOND PDZ DOMAIN OF SYNTENIN | | Descriptor: | OXYGEN ATOM, Syntenin 1 | | Authors: | Kang, B.S, Cooper, D.R, Devedjiev, Y, Derewenda, U, Derewenda, Z.S. | | Deposit date: | 2003-01-29 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Molecular roots of degenerate specificity in syntenin's PDZ2 domain: reassessment of the PDZ recognition paradigm.
Structure, 11, 2003
|
|
1NTF
 
 | | Crystal Structure of Cimex Nitrophorin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, salivary nitrophorin | | Authors: | Weichsel, A, Maes, E.M, Andersen, J.F, Valenzuela, J.G, Walker, F.A, Montfort, W.R. | | Deposit date: | 2003-01-29 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1NTG
 
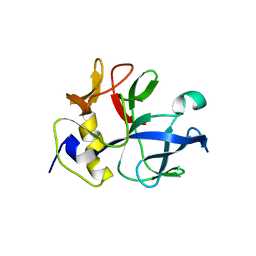 | |
1NTH
 
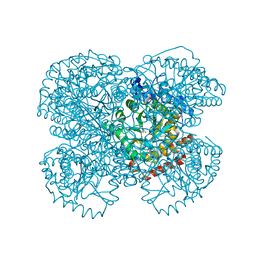 | | Crystal structure of the methanosarcina barkeri monomethylamine methyltransferase (MTMB) | | Descriptor: | Monomethylamine methyltransferase mtmB1 | | Authors: | Hao, B, Gong, W, Ferguson, T.K, James, C.M, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A new UAG-encoded residue in the structure of a methanogen methyltransferase
Science, 296, 2002
|
|
1NTI
 
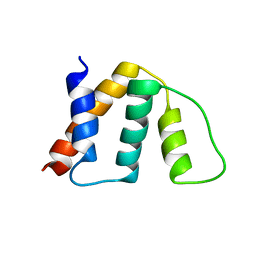 | |
1NTJ
 
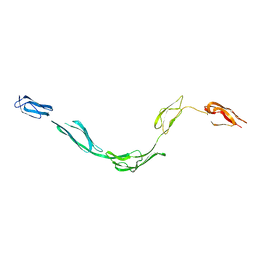 | | Model of rat Crry determined by solution scattering, curve fitting and homology modelling | | Descriptor: | complement receptor related protein | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The Extended Multidomain Solution Structures of the Complement Protein
Crry and its Chimeric Conjugate Crry-Ig by Scattering, Analytical
Ultracentrifugation and Constrained Modelling: Implications for Function and
Therapy
J.Mol.Biol., 329, 2003
|
|
1NTK
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Antimycin A1 | | Descriptor: | Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C.-A, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2016-03-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
1NTL
 
 | | Model of mouse Crry-Ig determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Complement component receptor 1-like protein,Ig gamma-1 chain C region secreted form | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The extended multidomain solution structures of the complement protein Crry
and its chimaeric conjugate Crry-Ig by scattering, analytical ultracentrifugation
and constrained modelling: implications for function and therapy
J.Mol.Biol., 329, 2003
|
|
1NTM
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 Complex at 2.4 Angstrom | | Descriptor: | Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
1NTN
 
 | | THE CRYSTAL STRUCTURE OF NEUROTOXIN-I FROM NAJA NAJA OXIANA AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | NEUROTOXIN I | | Authors: | Mikhailov, A.M, Nickitenko, A.V, Vainshtein, B.K, Betzel, C, Wilson, K. | | Deposit date: | 1994-09-26 | | Release date: | 1995-05-08 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-dimensional structure of neurotoxin-1 from Naja naja oxiana venom at 1.9 A resolution.
Febs Lett., 320, 1993
|
|
1NTO
 
 | | N249Y MUTANT OF ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-MONOCLINIC CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-01-30 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity.
Febs Lett., 539, 2003
|
|
