1L7N
 
 | | TRANSITION STATE ANALOGUE OF PHOSPHOSERINE PHOSPHATASE (ALUMINUM FLUORIDE COMPLEX) | | Descriptor: | ALUMINUM FLUORIDE, MAGNESIUM ION, PHOSPHOSERINE PHOSPHATASE, ... | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7O
 
 | | CRYSTAL STRUCTURE OF PHOSPHOSERINE PHOSPHATASE IN APO FORM | | Descriptor: | ACETIC ACID, PHOSPHOSERINE PHOSPHATASE, ZINC ION | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7P
 
 | | SUBSTRATE BOUND PHOSPHOSERINE PHOSPHATASE COMPLEX STRUCTURE | | Descriptor: | PHOSPHATE ION, PHOSPHOSERINE, PHOSPHOSERINE PHOSPHATASE | | Authors: | Wang, W, Cho, H.S, Kim, R, Jancarik, J, Yokota, H, Nguyen, H.H, Grigoriev, I.V, Wemmer, D.E, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-03-16 | | Release date: | 2002-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic "snapshots" of intermediate states.
J.Mol.Biol., 319, 2002
|
|
1L7Q
 
 | | Ser117Ala Mutant of Bacterial Cocaine Esterase cocE | | Descriptor: | BENZOIC ACID, cocaine esterase | | Authors: | Turner, J.M, Larsen, N.A, Basran, A, Barbas III, C.F, Bruce, N.C, Wilson, I.A, Lerner, R.A. | | Deposit date: | 2002-03-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Biochemical characterization and structural analysis of a highly proficient cocaine
esterase
Biochemistry, 41, 2002
|
|
1L7R
 
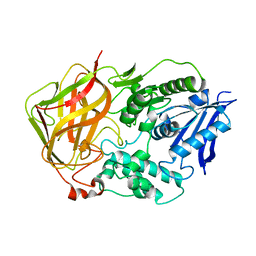 | | Tyr44Phe Mutant of Bacterial Cocaine Esterase cocE | | Descriptor: | cocaine esterase | | Authors: | Turner, J.M, Larsen, N.A, Basran, A, Barbas III, C.F, Bruce, N.C, Wilson, I.A, Lerner, R.A. | | Deposit date: | 2002-03-16 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Biochemical characterization and structural analysis of a highly proficient cocaine
esterase.
Biochemistry, 41, 2002
|
|
1L7T
 
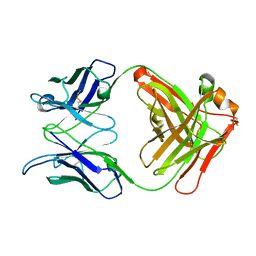 | | Crystal Structure Analysis of the anti-testosterone Fab fragment | | Descriptor: | anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2002-03-17 | | Release date: | 2002-10-02 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
1L7V
 
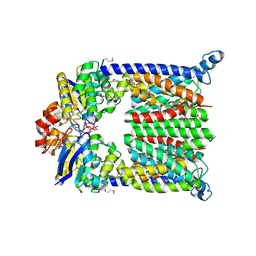 | | Bacterial ABC Transporter Involved in B12 Uptake | | Descriptor: | CYCLO-TETRAMETAVANADATE, VITAMIN B12 TRANSPORT SYSTEM PERMEASE PROTEIN BTUC, Vitamin B12 transport ATP-binding protein btuD | | Authors: | Locher, K.P, Lee, A.T, Rees, D.C. | | Deposit date: | 2002-03-18 | | Release date: | 2002-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism.
Science, 296, 2002
|
|
1L7X
 
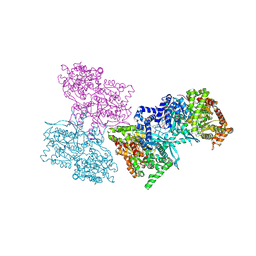 | | Human liver glycogen phosphorylase b complexed with caffeine, N-acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAFFEINE, Glycogen phosphorylase, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-18 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
1L7Y
 
 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
1L7Z
 
 | | Crystal structure of Ca2+/Calmodulin complexed with myristoylated CAP-23/NAP-22 peptide | | Descriptor: | CALCIUM ION, CALMODULIN, CAP-23/NAP-22, ... | | Authors: | Matsubara, M, Nakatsu, T, Yamauchi, E, Kato, H, Taniguchi, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-18 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a myristoylated CAP-23/NAP-22 N-terminal domain complexed with Ca2+/calmodulin
EMBO J., 23, 2004
|
|
1L80
 
 | |
1L81
 
 | |
1L82
 
 | |
1L83
 
 | |
1L84
 
 | |
1L85
 
 | |
1L86
 
 | |
1L87
 
 | |
1L88
 
 | |
1L89
 
 | |
1L8A
 
 | | E. COLI PYRUVATE DEHYDROGENASE | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component, THIAMINE DIPHOSPHATE | | Authors: | Furey, W, Arjunan, P. | | Deposit date: | 2002-03-19 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution.
Biochemistry, 41, 2002
|
|
1L8B
 
 | | Cocrystal Structure of the Messenger RNA 5' Cap-binding Protein (eIF4E) bound to 7-methylGpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E | | Authors: | Niedzwiecka, A, Marcotrigiano, J, Stepinski, J, Jankowska-Anyszka, M, Wyslouch-Cieszynska, A, Dadlez, M, Gingras, A.-C, Mak, P, Darzynkiewicz, E, Sonenberg, N. | | Deposit date: | 2002-03-19 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5' cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins.
J.Mol.Biol., 319, 2002
|
|
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L8D
 
 | | Rad50 coiled-coil Zn hook | | Descriptor: | CITRIC ACID, DNA double-strand break repair rad50 ATPase, MERCURY (II) ION, ... | | Authors: | Hopfner, K.P, Tainer, J.A. | | Deposit date: | 2002-03-20 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair.
Nature, 418, 2002
|
|
1L8F
 
 | |
