1JAW
 
 | | AMINOPEPTIDASE P FROM E. COLI LOW PH FORM | | Descriptor: | ACETATE ION, AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Wilce, M.C.J, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1JAX
 
 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) | | Descriptor: | MAGNESIUM ION, SODIUM ION, conserved hypothetical protein | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
1JAY
 
 | | Structure of Coenzyme F420H2:NADP+ Oxidoreductase (FNO) with its substrates bound | | Descriptor: | COENZYME F420, Coenzyme F420H2:NADP+ Oxidoreductase (FNO), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Warkentin, E, Mamat, B, Thauer, R, Ermler, U, Shima, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of F420H2:NADP+ oxidoreductase with and without its substrates bound.
EMBO J., 20, 2001
|
|
1JAZ
 
 | |
1JB0
 
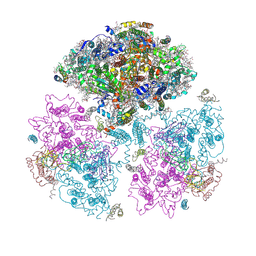 | | Crystal Structure of Photosystem I: a Photosynthetic Reaction Center and Core Antenna System from Cyanobacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Jordan, P, Fromme, P, Witt, H.T, Klukas, O, Saenger, W, Krauss, N. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-01 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional Structure of Cyanobacterial Photosystem I at 2.5 A Resolution
NATURE, 411, 2001
|
|
1JB1
 
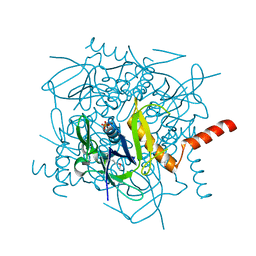 | | Lactobacillus casei HprK/P Bound to Phosphate | | Descriptor: | HPRK PROTEIN, PHOSPHATE ION | | Authors: | Fieulaine, S, Morera, S, Poncet, S, Monedero, V, Gueguen-Chaignon, V, Galinier, A, Janin, J, Deutscher, J, Nessler, S. | | Deposit date: | 2001-06-01 | | Release date: | 2001-08-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of HPr kinase: a bacterial protein kinase with a P-loop nucleotide-binding domain.
EMBO J., 20, 2001
|
|
1JB2
 
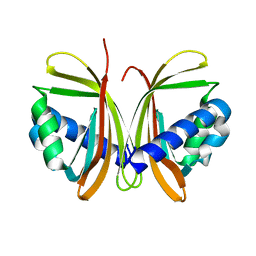 | | CRYSTAL STRUCTURE OF NTF2 M84E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JB3
 
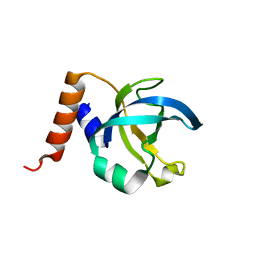 | |
1JB4
 
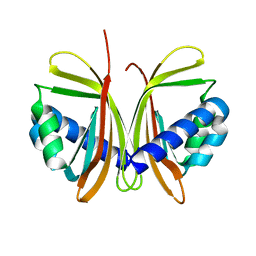 | | CRYSTAL STRUCTURE OF NTF2 M102E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JB5
 
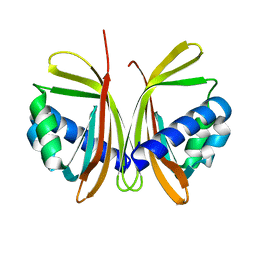 | | CRYSTAL STRUCTURE OF NTF2 M118E MUTANT | | Descriptor: | NUCLEAR TRANSPORT FACTOR 2 | | Authors: | Chaillan-Huntington, C, Butler, P.J, Huntington, J.A, Akin, D, Feldherr, C, Stewart, M. | | Deposit date: | 2001-06-01 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NTF2 monomer-dimer equilibrium.
J.Mol.Biol., 314, 2001
|
|
1JB6
 
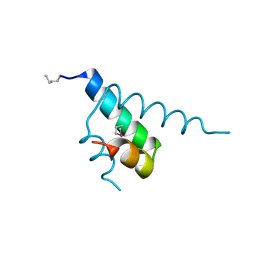 | |
1JB7
 
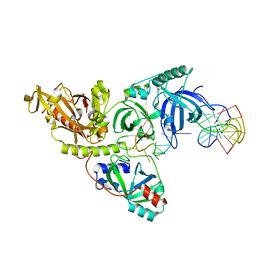 | |
1JB8
 
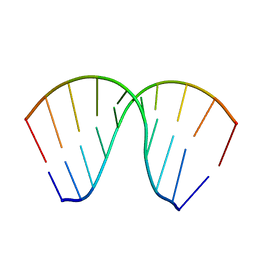 | | The Crystal Structure of an RNA/DNA Hybrid Reveals Novel Intermolecular Intercalation | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3' | | Authors: | Han, G.W, Kopka, M.L, Langs, D, Dickerson, R.E. | | Deposit date: | 2001-06-02 | | Release date: | 2003-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of an RNADNA hybrid reveals intermolecular
intercalation: Dimer formation by base-pair swapping
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1JB9
 
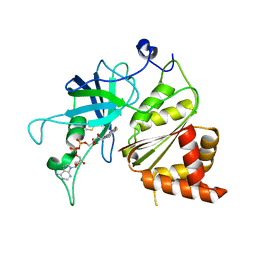 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
1JBA
 
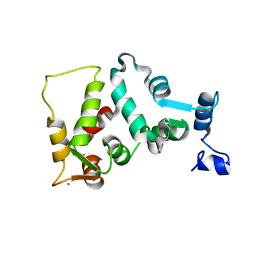 | | UNMYRISTOYLATED GCAP-2 WITH THREE CALCIUM IONS BOUND | | Descriptor: | CALCIUM ION, PROTEIN (GUANYLATE CYCLASE ACTIVATING PROTEIN 2) | | Authors: | Ames, J.B, Dizhoor, A.M, Ikura, M, Palczewski, K, Stryer, L. | | Deposit date: | 1999-04-03 | | Release date: | 1999-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of guanylyl cyclase activating protein-2, a calcium-sensitive modulator of photoreceptor guanylyl cyclases.
J.Biol.Chem., 274, 1999
|
|
1JBB
 
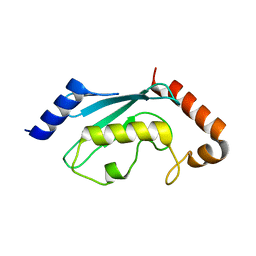 | | Ubiquitin Conjugating Enzyme, Ubc13 | | Descriptor: | ubiquitin conjugating enzyme E2-17.5 KDA | | Authors: | VanDemark, A.P, Hofmann, R.M, Tsui, C, Pickart, C.M, Wolberger, C. | | Deposit date: | 2001-06-03 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
1JBC
 
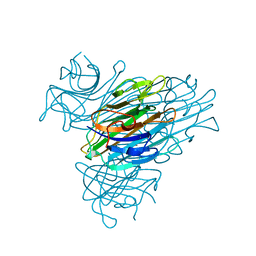 | | CONCANAVALIN A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Parkin, S, Rupp, B, Hope, H. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structure of concanavalin A at 120 K.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1JBD
 
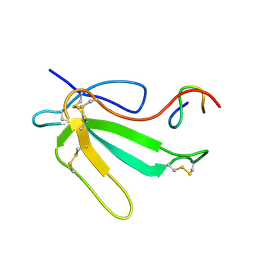 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1JBE
 
 | |
1JBF
 
 | |
1JBG
 
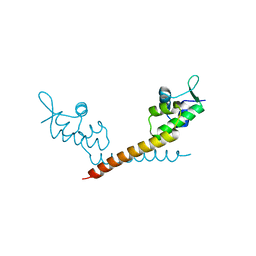 | | Crystal Structure of MtaN, the Bacillus subtilis Multidrug Transporter Activator, N-terminus | | Descriptor: | transcription activator of multidrug-efflux transporter genes mta | | Authors: | Godsey, M.H, Neyfakh, A.A, Brennan, R.G. | | Deposit date: | 2001-06-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of MtaN, a global multidrug transporter gene activator.
J.Biol.Chem., 276, 2001
|
|
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
1JBI
 
 | | NMR structure of the LCCL domain | | Descriptor: | cochlin | | Authors: | Liepinsh, E, Trexler, M, Kaikkonen, A, Weigelt, J, Banyai, L, Patthy, L, Otting, G. | | Deposit date: | 2001-06-05 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the LCCL domain and implications for DFNA9 deafness disorder.
EMBO J., 20, 2001
|
|
1JBJ
 
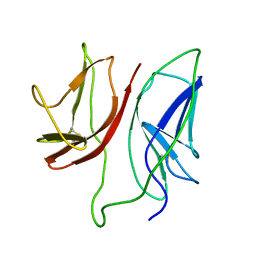 | | CD3 Epsilon and gamma Ectodomain Fragment Complex in Single-Chain Construct | | Descriptor: | CD3 Epsilon and gamma Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, K.S, Wagner, G, Reinherz, E.L. | | Deposit date: | 2001-06-05 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Mechanisms contributing to T cell receptor signaling and assembly revealed by the solution structure of an ectodomain fragment of the CD3 epsilon gamma heterodimer.
Cell(Cambridge,Mass.), 105, 2001
|
|
1JBK
 
 | |
