1HQY
 
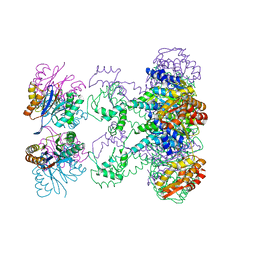 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-20 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
1HQZ
 
 | | Cofilin homology domain of a yeast actin-binding protein ABP1P | | Descriptor: | ACTIN-BINDING PROTEIN | | Authors: | Strokopytov, B.V, Fedorov, A.A, Mahoney, N, Drubin, D.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2000-12-20 | | Release date: | 2001-12-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1HR0
 
 | | CRYSTAL STRUCTURE OF INITIATION FACTOR IF1 BOUND TO THE 30S RIBOSOMAL SUBUNIT | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Carter, A.P, Clemons Jr, W.M, Brodersen, D.E, Morgan-Warren, R.J, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2000-12-20 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an initiation factor bound to the 30S ribosomal subunit.
Science, 291, 2001
|
|
1HR1
 
 | |
1HR2
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT P4-P6 DOMAIN (DELC209) OF TETRAHYMENA THEMOPHILA GROUP I INTRON. | | Descriptor: | MAGNESIUM ION, P4-P6 DELC209 MUTANT RNA RIBOZYME DOMAIN | | Authors: | Juneau, K, Podell, E.R, Harrington, D.J, Cech, T.R. | | Deposit date: | 2000-12-20 | | Release date: | 2001-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA--solvent interactions.
Structure, 9, 2001
|
|
1HR3
 
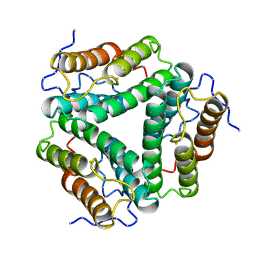 | |
1HR6
 
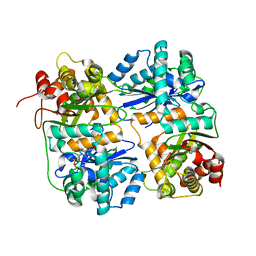 | | Yeast Mitochondrial Processing Peptidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR7
 
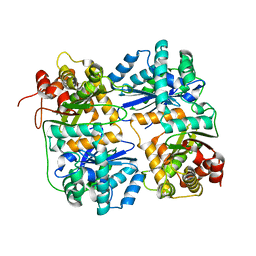 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant | | Descriptor: | MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, MITOCHONDRIAL PROCESSING PEPTIDASE BETA SUBUNIT, ZINC ION | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR8
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Cytochrome C Oxidase IV Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYTOCHROME C OXIDASE POLYPEPTIDE IV, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HR9
 
 | | Yeast Mitochondrial Processing Peptidase beta-E73Q Mutant Complexed with Malate Dehydrogenase Signal Peptide | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MALATE DEHYDROGENASE, MITOCHONDRIAL PROCESSING PEPTIDASE ALPHA SUBUNIT, ... | | Authors: | Taylor, A.B, Smith, B.S, Kitada, S, Kojima, K, Miyaura, H, Otwinowski, Z, Ito, A, Deisenhofer, J. | | Deposit date: | 2000-12-21 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences.
Structure, 9, 2001
|
|
1HRA
 
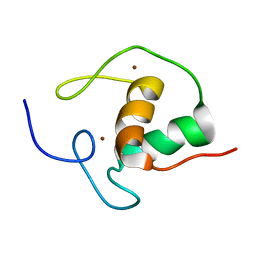 | | THE SOLUTION STRUCTURE OF THE HUMAN RETINOIC ACID RECEPTOR-BETA DNA-BINDING DOMAIN | | Descriptor: | RETINOIC ACID RECEPTOR, ZINC ION | | Authors: | Knegtel, R.M.A, Katahira, M, Schilthuis, J.G, Bonvin, A.M.J.J, Boelens, R, Eib, D, Van Der Saag, P.T, Kaptein, R. | | Deposit date: | 1993-07-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the human retinoic acid receptor-beta DNA-binding domain.
J.Biomol.NMR, 3, 1993
|
|
1HRB
 
 | |
1HRC
 
 | |
1HRD
 
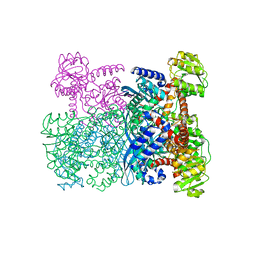 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Britton, K.L, Baker, P.J, Stillman, T.J, Rice, D.W. | | Deposit date: | 1996-04-03 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures.
Structure, 3, 1995
|
|
1HRE
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRF
 
 | | SOLUTION STRUCTURE OF THE EPIDERMAL GROWTH FACTOR-LIKE DOMAIN OF HEREGULIN-ALPHA, A LIGAND FOR P180ERB4 | | Descriptor: | HEREGULIN ALPHA | | Authors: | Nagata, K, Kohda, D, Hatanaka, H, Ichikawa, S, Inagaki, F. | | Deposit date: | 1994-07-21 | | Release date: | 1994-10-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the epidermal growth factor-like domain of heregulin-alpha, a ligand for p180erbB-4.
EMBO J., 13, 1994
|
|
1HRH
 
 | |
1HRI
 
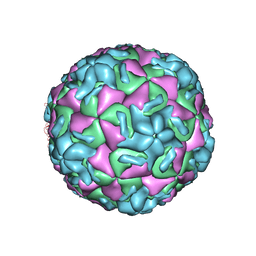 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
1HRJ
 
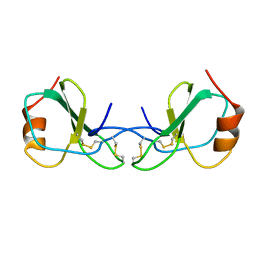 | | HUMAN RANTES, NMR, 13 STRUCTURES | | Descriptor: | HUMAN REGULATED UPON ACTIVATION NORMAL T-CELL EXPRESSED AND SECRETED | | Authors: | Chung, C, Cooke, R.M, Proudfoot, A.E.I, Wells, T.N.C. | | Deposit date: | 1995-08-18 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of RANTES.
Biochemistry, 34, 1995
|
|
1HRK
 
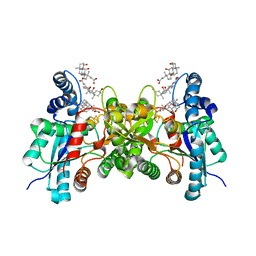 | | CRYSTAL STRUCTURE OF HUMAN FERROCHELATASE | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, FERROCHELATASE | | Authors: | Wu, C.K, Dailey, H.A, Rose, J.P, Burden, A, Sellers, V.M, Wang, B.-C. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1HRL
 
 | |
1HRM
 
 | |
1HRN
 
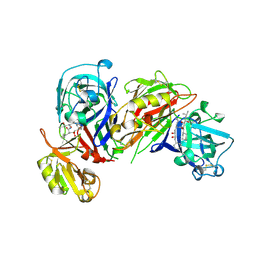 | | HIGH RESOLUTION CRYSTAL STRUCTURES OF RECOMBINANT HUMAN RENIN IN COMPLEX WITH POLYHYDROXYMONOAMIDE INHIBITORS | | Descriptor: | (2R,4S,5S)-N-[(2S,3R,4S)-1-cyclohexyl-3,4-dihydroxy-6-methylheptan-2-yl]-2-(cyclopropylmethyl)-4,5-dihydroxy-6-phenylhexanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, RENIN | | Authors: | Tong, L, Anderson, P.C. | | Deposit date: | 1995-03-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution crystal structures of recombinant human renin in complex with polyhydroxymonoamide inhibitors.
J.Mol.Biol., 250, 1995
|
|
1HRO
 
 | |
1HRP
 
 | | CRYSTAL STRUCTURE OF HUMAN CHORIONIC GONADOTROPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN CHORIONIC GONADOTROPIN | | Authors: | Lapthorn, A.J, Harris, D.C, Isaacs, N.W. | | Deposit date: | 1994-08-15 | | Release date: | 1994-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human chorionic gonadotropin.
Nature, 369, 1994
|
|
