168L
 
 | |
169D
 
 | | THE SOLUTION STRUCTURE OF THE R(GCG)D(TATACCC):D(GGGTATACGC) OKAZAKI FRAGMENT CONTAINS TWO DISTINCT DUPLEX MORPHOLOGIES CONNECTED BY A JUNCTION | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*AP*TP*AP*CP*GP*C)-3'), DNA/RNA (5'-R(*GP*CP*G)-D(P*TP*AP*TP*AP*CP*CP*C)-3') | | Authors: | Salazar, M, Fedoroff, O.Y, Zhu, L, Reid, B.R. | | Deposit date: | 1994-04-11 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the r(gcg)d(TATACCC):d(GGGTATACGC) Okazaki fragment contains two distinct duplex morphologies connected by a junction.
J.Mol.Biol., 241, 1994
|
|
169L
 
 | |
16GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 APO FORM 3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Oakley, A.J, Lo Bello, M, Ricci, G, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-30 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evidence for an induced-fit mechanism operating in pi class glutathione transferases.
Biochemistry, 37, 1998
|
|
16PK
 
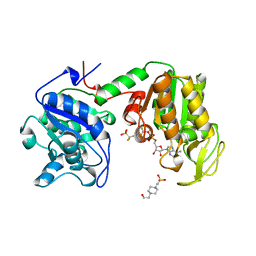 | | PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI BISUBSTRATE ANALOG | | Descriptor: | 1,1,5,5-TETRAFLUOROPHOSPHOPENTYLPHOSPHONIC ACID ADENYLATE ESTER, 3-PHOSPHOGLYCERATE KINASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | | Authors: | Bernstein, B.E, Bressi, J, Blackburn, M, Gelb, M, Hol, W.G.J. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bisubstrate analog induces unexpected conformational changes in phosphoglycerate kinase from Trypanosoma brucei.
J.Mol.Biol., 279, 1998
|
|
16VP
 
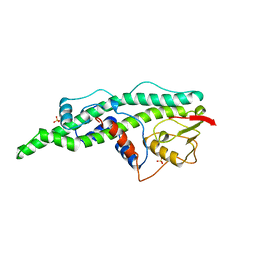 | | CONSERVED CORE OF THE HERPES SIMPLEX VIRUS TRANSCRIPTIONAL REGULATORY PROTEIN VP16 | | Descriptor: | PROTEIN (VP16, VMW65, ATIF), ... | | Authors: | Liu, Y, Gong, W, Huang, C.C, Herr, W, Cheng, X. | | Deposit date: | 1999-02-11 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16.
Genes Dev., 13, 1999
|
|
170D
 
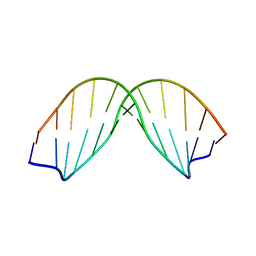 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA/RNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*T)-R(P*CAR)-D(P*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
170L
 
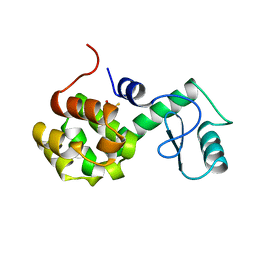 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L.H, Wozniak, A, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
171D
 
 | | SOLUTION STRUCTURE OF A DNA DODECAMER CONTAINING THE ANTI-NEOPLASTIC AGENT ARABINOSYLCYTOSINE: COMBINED USE OF NMR, RESTRAINED MOLECULAR DYNAMICS AND FULL RELAXATION MATRIX REFINEMENT | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Schweitzer, B.I, Mikita, T, Kellogg, G.W, Gardner, K.H, Beardsley, G.P. | | Deposit date: | 1994-03-14 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinosylcytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement.
Biochemistry, 33, 1994
|
|
171L
 
 | |
172D
 
 | |
172L
 
 | |
173D
 
 | | MULTIPLE BINDING MODES OF ANTICANCER DRUG ACTINOMYCIN D: X-RAY, MOLECULAR MODELING, AND SPECTROSCOPIC STUDIES OF D(GAAGCTTC)2-ACTINOMYCIN D COMPLEXES AND ITS HOST DNA | | Descriptor: | ACTINOMYCIN D, DNA (5'-D(*GP*AP*AP*GP*CP*TP*TP*C)-3') | | Authors: | Kamitori, S, Takusagawa, F. | | Deposit date: | 1994-04-18 | | Release date: | 1994-10-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multiple Binding Modes of Anticancer Drug Actinomycin D: X-Ray, Molecular Modeling, and Spectroscopic Studies of D(Gaagcttc)2-Actinomycin D Complexes and its Host DNA
J.Am.Chem.Soc., 116, 1994
|
|
173L
 
 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Xiong, X.-P, Zhang, X.-J, Sun, D, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
174L
 
 | |
175D
 
 | |
175L
 
 | |
176D
 
 | |
176L
 
 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L, Dubose, R, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
177D
 
 | |
177L
 
 | |
178D
 
 | | CRYSTAL STRUCTURE OF A DNA DUPLEX CONTAINING 8-HYDROXYDEOXYGUANINE.ADENINE BASE-PAIRS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*(8OG)P*GP*CP*G)-3') | | Authors: | McAuley-Hecht, K.E, Leonard, G.A, Gibson, N.J, Thomson, J.B, Watson, W.P, Hunter, W.N, Brown, T. | | Deposit date: | 1994-05-19 | | Release date: | 1994-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs.
Biochemistry, 33, 1994
|
|
178L
 
 | | Protein flexibility and adaptability seen in 25 crystal forms of T4 LYSOZYME | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Matsumura, M, Weaver, L, Zhang, X.-J, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
179D
 
 | |
17GS
 
 | | GLUTATHIONE S-TRANSFERASE P1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-07 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glutathione S-transferase P1-1
To be published
|
|
