1B28
 
 | | ARC REPRESSOR MYL MUTANT FROM SALMONELLA BACTERIOPHAGE P22 | | Descriptor: | PROTEIN (REGULATORY PROTEIN ARC) | | Authors: | Rietveld, A.W.M, Nooren, I.M.A, Sauer, R.T, Kaptein, R, Boelens, R. | | Deposit date: | 1998-12-05 | | Release date: | 1999-11-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of an Arc repressor mutant reveal premelting conformational changes related to DNA binding.
Biochemistry, 38, 1999
|
|
1B2A
 
 | |
1B2B
 
 | |
1B2C
 
 | |
1B2D
 
 | |
1B2E
 
 | |
1B2F
 
 | |
1B2G
 
 | |
1B2H
 
 | |
1B2I
 
 | |
1B2J
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2K
 
 | | Structural effects of monovalent anions on polymorphic lysozyme crystals | | Descriptor: | IODIDE ION, PROTEIN (LYSOZYME) | | Authors: | Vaney, M.C, Broutin, I, Ries-Kautt, M, Ducruix, A. | | Deposit date: | 1998-11-26 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural effects of monovalent anions on polymorphic lysozyme crystals.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1B2L
 
 | | ALCOHOL DEHYDROGENASE FROM DROSOPHILA LEBANONENSIS: TERNARY COMPLEX WITH NAD-CYCLOHEXANONE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ALCOHOL DEHYDROGENASE, CALCIUM ION, ... | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
1B2M
 
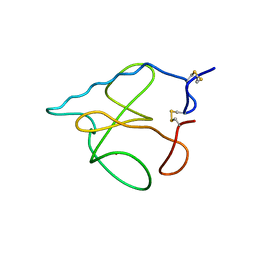 | | THREE-DIMENSIONAL STRUCTURE OF RIBONULCEASE T1 COMPLEXED WITH AN ISOSTERIC PHOSPHONATE ANALOGUE OF GPU: ALTERNATE SUBSTRATE BINDING MODES AND CATALYSIS. | | Descriptor: | 5'-R(*GP*(U34))-3', RIBONUCLEASE T1 | | Authors: | Arni, R.K, Watanabe, L, Ward, R.J, Kreitman, R.J, Kumar, K, Walz Jr, F.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of ribonuclease T1 complexed with an isosteric phosphonate substrate analogue of GpU: alternate substrate binding modes and catalysis.
Biochemistry, 38, 1999
|
|
1B2O
 
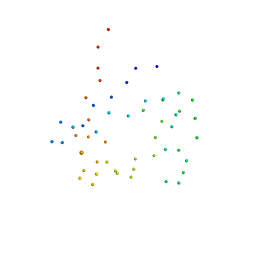 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2P
 
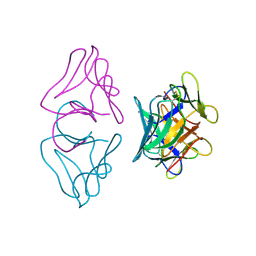 | | NATIVE MANNOSE-SPECIFIC BULB LECTIN FROM SCILLA CAMPANULATA (BLUEBELL) AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (LECTIN) | | Authors: | Wood, S.D, Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, Peumans, W.J, Van Damme, E.J.M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the native (unligated) mannose-specific bulb lectin from Scilla campanulata (bluebell) at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2R
 
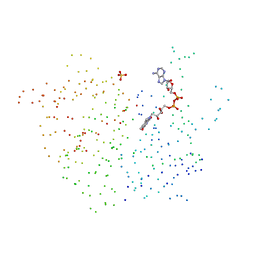 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: E 301 A) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN-NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-11-27 | | Release date: | 1999-12-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the catalytic role of Glu301 in Anabaena PCC 7119 ferredoxin-NADP+ reductase revealed by x-ray crystallography.
Proteins, 38, 2000
|
|
1B2S
 
 | |
1B2T
 
 | |
1B2U
 
 | |
1B2V
 
 | | HEME-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, PROTEIN (HEME-BINDING PROTEIN A), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi, N, Lecroisey, A, Wandersma, N.C, Czjzek, M. | | Deposit date: | 1998-12-01 | | Release date: | 1999-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of HasA, a hemophore secreted by Serratia marcescens.
Nat.Struct.Biol., 6, 1999
|
|
1B2W
 
 | | COMPARISON OF THE THREE-DIMENSIONAL STRUCTURES OF A HUMANIZED AND A CHIMERIC FAB OF AN ANTI-GAMMA-INTERFERON ANTIBODY | | Descriptor: | PROTEIN (ANTIBODY (HEAVY CHAIN)), PROTEIN (ANTIBODY (LIGHT CHAIN)) | | Authors: | Fan, Z, Shan, L, Goldsteen, B.Z, Guddat, L.W, Thakur, A, Landolfi, N.F, Co, M.S, Vasquez, M, Queen, C, Ramsland, P.A, Edmundson, A.B. | | Deposit date: | 1998-12-01 | | Release date: | 1999-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of the three-dimensional structures of a humanized and a chimeric Fab of an anti-gamma-interferon antibody.
J.Mol.Recog., 12, 1999
|
|
1B2X
 
 | | BARNASE WILDTYPE STRUCTURE AT PH 7.5 FROM A CRYO_COOLED CRYSTAL AT 100K | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Harrison, P, Vaughan, C.K, Buckle, A.M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1B2Y
 
 | |
1B2Z
 
 | | DELETION OF A BURIED SALT BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
