7TMS
 
 | | V-ATPase from Saccharomyces cerevisiae, State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMR
 
 | | V-ATPase from Saccharomyces cerevisiae, State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TMT
 
 | | V-ATPase from Saccharomyces cerevisiae, State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, H(+)-transporting two-sector ATPase, V-ATPase subunit E, ... | | Authors: | Vasanthakumar, T, Keon, K.A, Bueler, S.A, Jaskolka, M.C, Rubinstein, J.L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-04-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Coordinated conformational changes in the V 1 complex during V-ATPase reversible dissociation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TAP
 
 | | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | | Descriptor: | Archazolid A, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7TAO
 
 | | Cryo-EM structure of bafilomycin A1 bound to yeast VO V-ATPase | | Descriptor: | (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7U8O
 
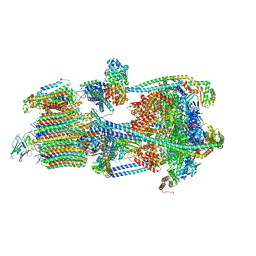 | | Structure of porcine V-ATPase with mEAK7 and SidK, Rotary state 2 | | Descriptor: | ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, Bacterial effector protein SidK, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8P
 
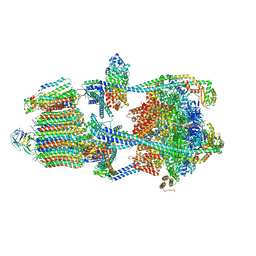 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8Q
 
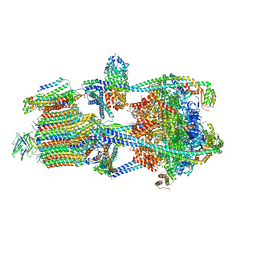 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8R
 
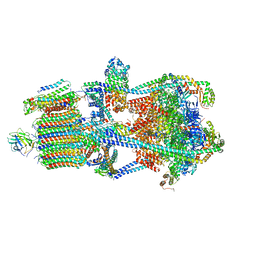 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U4T
 
 | | Human V-ATPase in state 2 with SidK and mEAK-7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MTOR-associated protein MEAK7, ... | | Authors: | Wang, L, Fu, T.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-08-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Identification of mEAK-7 as a human V-ATPase regulator via cryo-EM data mining.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5VOX
 
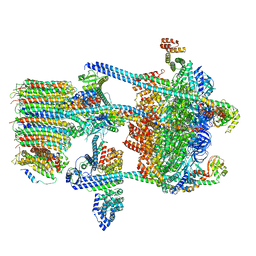 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
4RND
 
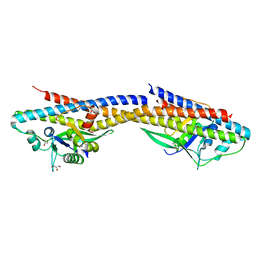 | |
4IX9
 
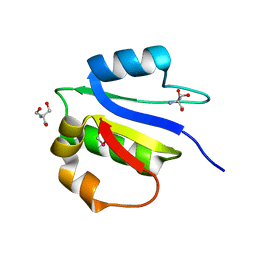 | | Crystal structure of subunit F of V-ATPase from S. cerevisiae | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, V-type proton ATPase subunit F | | Authors: | Basak, S, Balakrishna, A.M, Manimekalai, M.S.S, Gruber, G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal and NMR structures give insights into the role and dynamics of subunit F of the eukaryotic V-ATPase from Saccharomyces cerevisiae
J.Biol.Chem., 288, 2013
|
|
2LX4
 
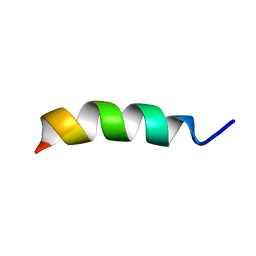 | |
1U7L
 
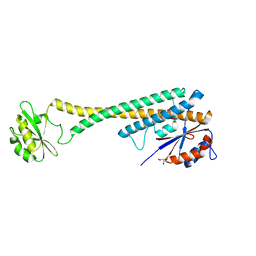 | |
2NVJ
 
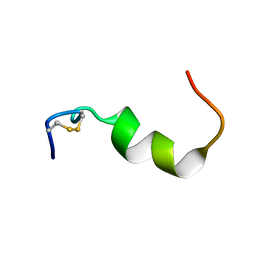 | | NMR structures of transmembrane segment from subunit a from the yeast proton V-ATPase | | Descriptor: | 25mer peptide from Vacuolar ATP synthase subunit a, vacuolar isoform | | Authors: | Hemminga, M.A, van Mierlo, C.P, Wechselberger, R, de Jong, E.R, Duarte, A.M. | | Deposit date: | 2006-11-13 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Segment TM7 from the cytoplasmic hemi-channel from V(O)-H(+)-V-ATPase includes a flexible region that has a potential role in proton translocation
Biochim.Biophys.Acta, 1768, 2007
|
|
5TJ5
 
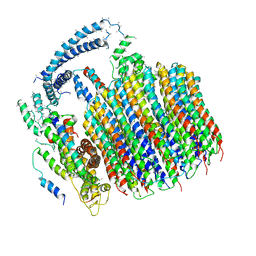 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | Descriptor: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2016-10-03 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
5I1M
 
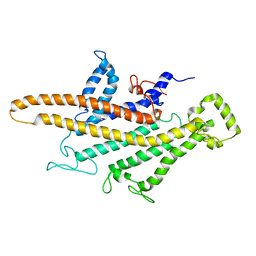 | | Yeast V-ATPase average of densities, a subunit segment | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Schep, D.G, Zhao, J, Rubinstein, J.L. | | Deposit date: | 2016-02-05 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RPW
 
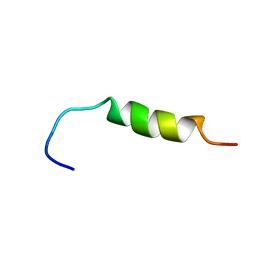 | | Structure of a peptide derived from H+-V-ATPase subunit a | | Descriptor: | 25 meric peptide from V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Vermeer, L.S, Reat, V, Hemminga, M.A, Milon, A. | | Deposit date: | 2008-11-08 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a peptide derived from H(+)-V-ATPase subunit a
Biochim.Biophys.Acta, 1788, 2009
|
|
5VOY
 
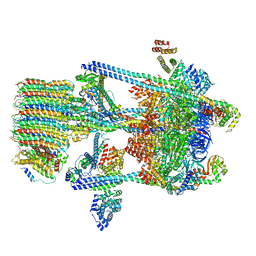 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 2) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VOZ
 
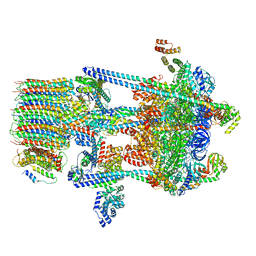 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 3) | | Descriptor: | Uncharacterized protein, V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
6WM2
 
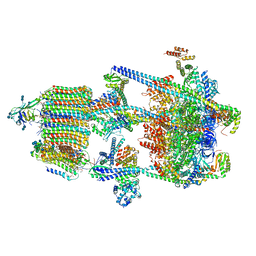 | | Human V-ATPase in state 1 with SidK and ADP | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM3
 
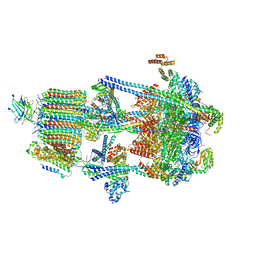 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6O7X
 
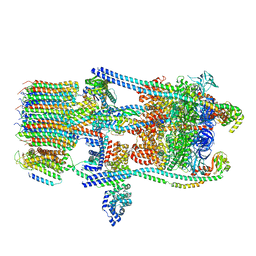 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6O7V
 
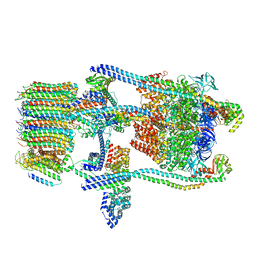 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
