7ZXZ
 
 | | dithiol-ligand bound to streptavidin | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(1~{R},3~{S})-3-[3,5-bis(sulfanylmethyl)phenyl]-2,4-bis(oxidanylidene)cyclopentyl]pentanamide, DI(HYDROXYETHYL)ETHER, Streptavidin | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | dithiol-ligand bound to streptavidin
To Be Published
|
|
2O88
 
 | |
4J9C
 
 | |
4J9G
 
 | |
4J9H
 
 | |
4J9I
 
 | |
2KBT
 
 | | Attachment of an NMR-invisible solubility enhancement tag (INSET) using a sortase-mediated protein ligation method | | Descriptor: | Proto-oncogene vav,Immunoglobulin G-binding protein G | | Authors: | Kumeta, H, Kobashigawa, Y, Ogura, K, Inagaki, F. | | Deposit date: | 2008-12-07 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method
J.Biomol.Nmr, 43, 2009
|
|
1IGA
 
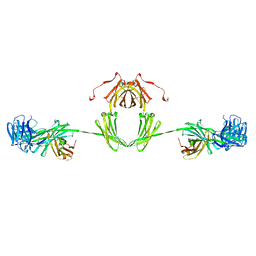 | |
3EG1
 
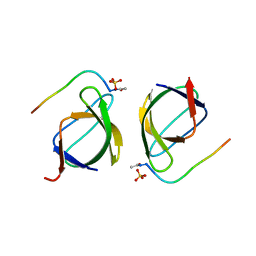 | |
5U2P
 
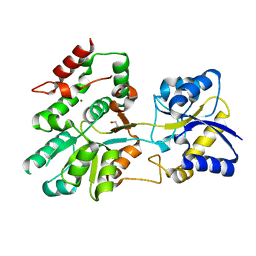 | | The crystal structure of Tp0737 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Brautigam, C.A, Deka, R.K, Tomchick, D.R, Norgard, M.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Functional clues from the crystal structure of an orphan periplasmic ligand-binding protein from Treponema pallidum.
Protein Sci., 26, 2017
|
|
1DJS
 
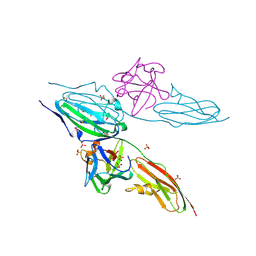 | |
7KV0
 
 | | Crystallographic structure of Paenibacillus xylanivorans GH11 | | Descriptor: | 1,2-ETHANEDIOL, Endo-1,4-beta-xylanase | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and molecular dynamics investigations of ligand stabilization via secondary binding site interactions in Paenibacillus xylanivorans GH11 xylanase.
Comput Struct Biotechnol J, 19, 2021
|
|
6BGD
 
 | | The crystal structure of the W145A variant of TpMglB-2 (Tp0684) with bound ligand | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/galactose-binding lipoprotein | | Authors: | Brautigam, C.A, Norgard, M.V, Deka, R.K. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structures of MglB-2 (TP0684), a topologically variant d-glucose-binding protein from Treponema pallidum, reveal a ligand-induced conformational change.
Protein Sci., 27, 2018
|
|
2CIK
 
 | | Insights Into Crossreactivity in Human Allorecognition: The Structure of HLA-B35011 Presenting an Epitope derived from Cytochrome P450. | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN B-35 ALPHA CHAIN, ... | | Authors: | Hourigan, C.S, Harkiolaki, M, Peterson, N.A, Bell, J.I, Jones, E.Y, O'Callaghan, C.A. | | Deposit date: | 2006-03-22 | | Release date: | 2006-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structure of the Human Allo-Ligand Hla-B3501 in Complex with a Cytochrome P450 Peptide: Steric Hindrance Influences Tcr Allo-Recognition.
Eur.J.Immunol., 36, 2006
|
|
1CS6
 
 | | N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN | | Descriptor: | AXONIN-1, GLYCEROL | | Authors: | Freigang, J, Proba, K, Diederichs, K, Sonderegger, P, Welte, W. | | Deposit date: | 1999-08-17 | | Release date: | 2000-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Cell(Cambridge,Mass.), 101, 2000
|
|
1AVN
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH THE HISTAMINE ACTIVATOR | | Descriptor: | AZIDE ION, CARBONIC ANHYDRASE II, HISTAMINE, ... | | Authors: | Briganti, F, Mangani, S, Orioli, P, Scozzafava, A, Vernaglione, G, Supuran, C.T. | | Deposit date: | 1997-09-17 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbonic anhydrase activators: X-ray crystallographic and spectroscopic investigations for the interaction of isozymes I and II with histamine.
Biochemistry, 36, 1997
|
|
8CI0
 
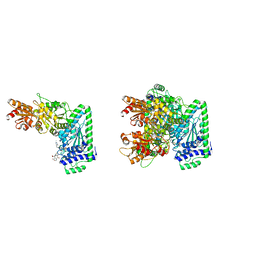 | | Maize Transketolase in complex with TPP and hydrolyzed (+)-Cornexistin | | Descriptor: | (1~{Z},3~{R},4~{S},7~{S},8~{Z})-8-ethylidene-4,7-bis(oxidanyl)-5-oxidanylidene-3-propyl-cyclononene-1,2-dicarboxylic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, MAGNESIUM ION, ... | | Authors: | Freigang, J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Investigations into Simplified Analogues of the Herbicidal Natural Product (+)-Cornexistin.
Chemistry, 29, 2023
|
|
6JQ4
 
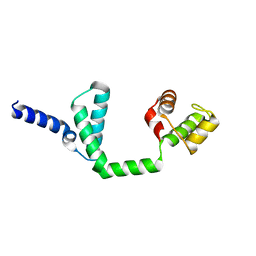 | | HIGA Escherichia coli-K12 | | Descriptor: | Antitoxin HigA | | Authors: | She, Z, Xu, B.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes of antitoxin HigA from Escherichia coli str. K-12 upon binding of its cognate toxin HigB reveal a new regulation mechanism in toxin-antitoxin systems.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
1JV0
 
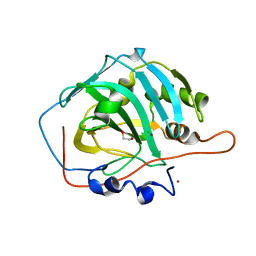 | | THE CRYSTAL STRUCTURE OF THE ZINC(II) ADDUCT OF THE CAI MICHIGAN 1 VARIANT | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, CHLORIDE ION, ... | | Authors: | Briganti, F, Ferraroni, M, Chegwidden, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-08-28 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination
Biochemistry, 41, 2002
|
|
1J9W
 
 | | Solution Structure of the CAI Michigan 1 Variant | | Descriptor: | 1,2-ETHANEDIOL, CARBONIC ANHYDRASE I, ZINC ION | | Authors: | Briganti, F, Ferraroni, M, Chedwiggen, W.R, Scozzafava, A, Supuran, C.T, Tilli, S. | | Deposit date: | 2001-05-29 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a zinc-activated variant of human carbonic anhydrase I, CA I Michigan 1: evidence for a second zinc binding site involving arginine coordination.
Biochemistry, 41, 2002
|
|
8TWZ
 
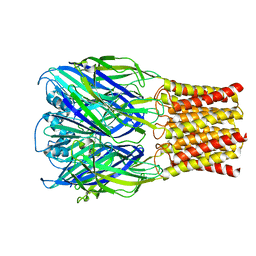 | | ELIC with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2023-08-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F35
 
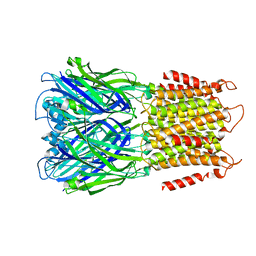 | | Apo ELIC in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F33
 
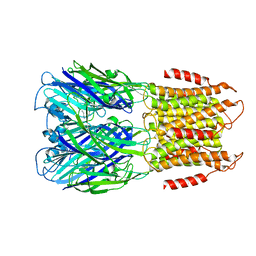 | | ELIC with Propylamine in saposin nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F34
 
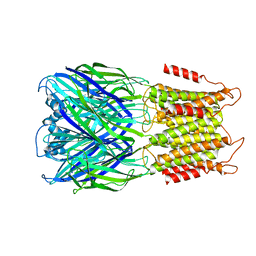 | | ELIC with Propylamine in spMSP1D1 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
8F32
 
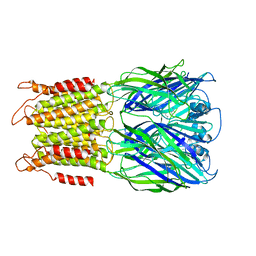 | | ELIC with Propylamine in SMA nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
