1UMH
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | F-box only protein 2, NICKEL (II) ION | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1UMJ
 
 | | Crystal structure of Pyrococcus horikoshii CutA in the presence of 3M guanidine hydrochloride | | Descriptor: | GUANIDINE, periplasmic divalent cation tolerance protein CutA | | Authors: | Tanaka, Y, Tsumoto, K, Yasutake, Y, Sakai, N, Yao, M, Tanaka, I, Kumagai, I. | | Deposit date: | 2003-10-02 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evidence for guanidine-protein side chain interactions: crystal structure of CutA from Pyrococcus horikoshii in 3M guanidine hydrochloride
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1UMK
 
 | | The Structure of Human Erythrocyte NADH-cytochrome b5 Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bando, S, Takano, T, Yubisui, T, Shirabe, K, Takeshita, M, Horii, C, Nakagawa, A. | | Deposit date: | 2003-10-03 | | Release date: | 2004-11-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human erythrocyte NADH-cytochrome b5 reductase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UML
 
 | | Crystal structure of adenosine deaminase complexed with a potent inhibitor FR233624 | | Descriptor: | 1-((1R)-1-(HYDROXYMETHYL)-3-{6-[(3-PHENYLPROPANOYL)AMINO]-1H-INDOL-1-YL}PROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-10-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1UMN
 
 | | Crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kauko, A, Haataja, S, Pulliainen, A, Finne, J, Papageorgiou, A.C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation.
J. Mol. Biol., 338, 2004
|
|
1UMO
 
 | | The crystal structure of cytoglobin: the fourth globin type discovered in man | | Descriptor: | CYTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | de Sanctis, D, Dewilde, S, Pesce, A, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-08-26 | | Release date: | 2004-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination.
J.Mol.Biol., 336, 2004
|
|
1UMP
 
 | |
1UMQ
 
 | |
1UMR
 
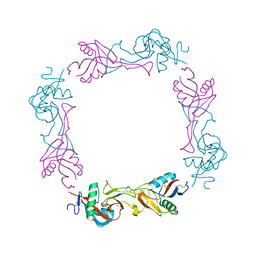 | | Crystal structure of the platelet activator convulxin, a disulfide linked a4b4 cyclic tetramer from the venom of Crotalus durissus terrificus | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Murakami, M.T, Zela, S.P, Gava, L.M, Michelan-Duarte, S, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Platelet Activator Convulxin, a Disulfide Linked A4B4 Cyclic Tetramer from the Venom of Crotalus Durissus Terrificus
Biochem.Biophys.Res.Commun., 310, 2003
|
|
1UMS
 
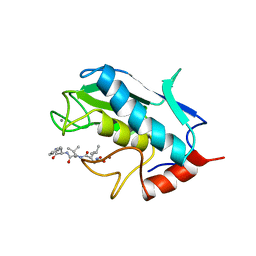 | | STROMELYSIN-1 CATALYTIC DOMAIN WITH HYDROPHOBIC INHIBITOR BOUND, PH 7.0, 32OC, 20 MM CACL2, 15% ACETONITRILE; NMR ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMT
 
 | | Stromelysin-1 catalytic domain with hydrophobic inhibitor bound, ph 7.0, 32oc, 20 mm cacl2, 15% acetonitrile; nmr average of 20 structures minimized with restraints | | Descriptor: | CALCIUM ION, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-L-leucyl-L-phenylalaninamide, STROMELYSIN-1, ... | | Authors: | Van Doren, S.R, Kurochkin, A.V, Hu, W, Zuiderweg, E.R.P. | | Deposit date: | 1995-10-31 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human stromelysin complexed with a hydrophobic inhibitor.
Protein Sci., 4, 1995
|
|
1UMU
 
 | |
1UMV
 
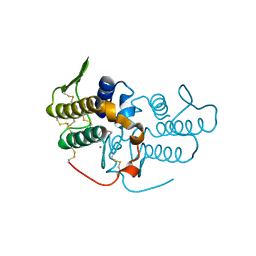 | | Crystal structure of an acidic, non-myotoxic phospholipase A2 from the venom of Bothrops jararacussu | | Descriptor: | CALCIUM ION, HYPOTENSIVE PHOSPHOLIPASE A2 | | Authors: | Murakami, M.T, Watanabe, L, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of an Acidic Platelet Aggregation Inhibitor and Hypotensive Phospholipase A(2) in the Monomeric and Dimeric States: Insights Into its Oligomeric State
Biochem.Biophys.Res.Commun., 323, 2004
|
|
1UMW
 
 | | Structure of a human Plk1 Polo-box domain/phosphopeptide complex | | Descriptor: | PEPTIDE, SERINE/THREONINE-PROTEIN KINASE PLK | | Authors: | Rellos, P, Elia, A, Yaffe, M.B, Smerdon, S.J. | | Deposit date: | 2003-08-29 | | Release date: | 2003-10-16 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Molecular Basis for Phosphodependent Substrate Targeting and Regulation of Plks by the Polo-Box Domain
Cell(Cambridge,Mass.), 115, 2003
|
|
1UMX
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ARG M267 REPLACED WITH LEU (CHAIN M, R267L) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, FE (III) ION, ... | | Authors: | Fyfe, P.K, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2003-09-02 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre.
Biochim.Biophys.Acta, 1608, 2004
|
|
1UMY
 
 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
1UMZ
 
 | | Xyloglucan endotransglycosylase in complex with the xyloglucan nonasaccharide XLLG. | | Descriptor: | XYLOGLUCAN ENDOTRANSGLYCOSYLASE, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A.M, Henriksson, H, Denman, S.E, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1UN0
 
 | | Crystal Structure of Yeast Karyopherin (Importin) alpha in complex with a Nup2p N-terminal fragment | | Descriptor: | IMPORTIN ALPHA SUBUNIT, NUCLEOPORIN NUP2 | | Authors: | Matsuura, Y, Lange, A, Harreman, M.T, Corbett, A.H, Stewart, M. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Nup2P Function in Cargo Release and Karyopherin Recycling in Nuclear Import
Embo J., 22, 2003
|
|
1UN1
 
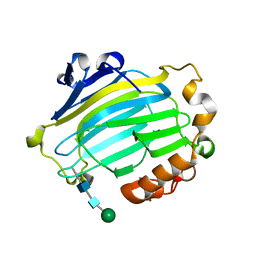 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1UN2
 
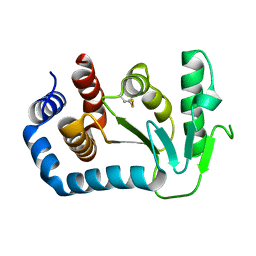 | | Crystal structure of circularly permuted CPDSBA_Q100T99: Preserved Global Fold and Local Structural Adjustments | | Descriptor: | THIOL-DISULFIDE INTERCHANGE PROTEIN | | Authors: | Manjasetty, B.A, Hennecke, J, Glockshuber, R, Heinemann, U. | | Deposit date: | 2003-09-03 | | Release date: | 2003-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Circularly Permuted Dsba(Q100T99): Preserved Global Fold and Local Structural Adjustments
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UN3
 
 | |
1UN4
 
 | |
1UN5
 
 | | ARH-II, AN ANGIOGENIN/RNASE A CHIMERA | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2004-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Studies on Structural Features that Determine the Enzymatic Specificity and Potency of Human Angiogenin: Thr44, Thr80 and Residues 38-41
Biochemistry, 43, 2004
|
|
1UN6
 
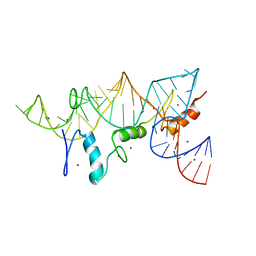 | | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | | Descriptor: | 5S RIBOSOMAL RNA, MAGNESIUM ION, TRANSCRIPTION FACTOR IIIA, ... | | Authors: | Lu, D, Searles, M.A, Klug, A. | | Deposit date: | 2003-09-04 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Zinc-Finger-RNA Complex Reveals Two Modes of Molecular Recognition
Nature, 426, 2003
|
|
