2KDN
 
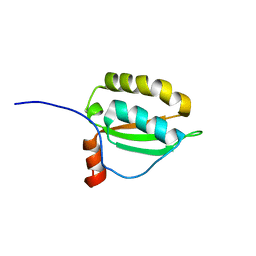 | | Solution structure of PFE0790c, a putative bolA-like protein from the protozoan parasite Plasmodium falciparum. | | Descriptor: | Putative uncharacterized protein PFE0790c | | Authors: | Buchko, G.W, Yee, A, Semesi, A, Hui, R, Arrowsmith, C.H, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-01-12 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution-state NMR structure of the putative morphogene protein BolA (PFE0790c) from Plasmodium falciparum.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2RNA
 
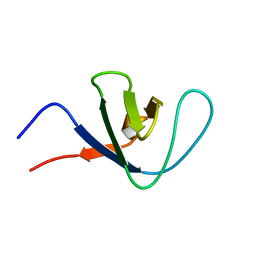 | | Itk SH3 average minimized | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Severin, A.J. | | Deposit date: | 2007-12-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure note: murine Itk SH3 domain
To be Published
|
|
2RUO
 
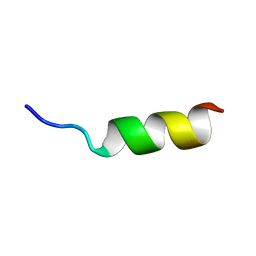 | | Solution Structure of Internal Fusion Peptide | | Descriptor: | UNP residues 873-888 of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
2RUM
 
 | | Solution structure of Fusion peptide | | Descriptor: | Fusion peptide of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-05 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
2RUN
 
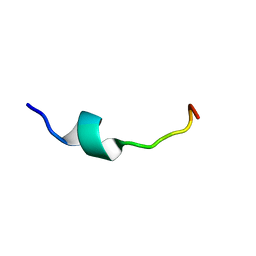 | | Solution Structure of Pre Transmembrane domain | | Descriptor: | Pre-transmembrane domain of Spike glycoprotein | | Authors: | Mahajan, M, Bhattacharjya, S. | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structures and Localization of Potential Fusion Peptides and Pre-transmembrane Region of SARS-CoV: Implications in Membrane Fusion
To be Published
|
|
1QXF
 
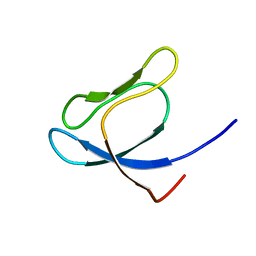 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S27E FROM ARCHAEOGLOBUS FULGIDUS: GR2, A NESG TARGET PROTEIN | | Descriptor: | 30S RIBOSOMAL PROTEIN S27E | | Authors: | Herve Du Penhoat, C, Atreya, H.S, Shen, Y, Liu, G, Acton, T.B, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the 30S ribosomal protein S27e encoded in gene RS27_ARCFU of Archaeoglobus fulgidis reveals a novel protein fold
Protein Sci., 13, 2004
|
|
1DGV
 
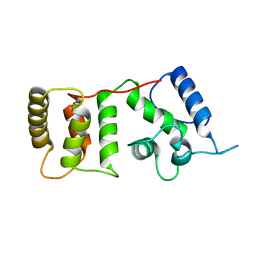 | |
1DGU
 
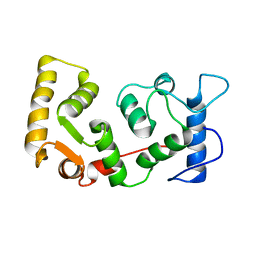 | |
1C2T
 
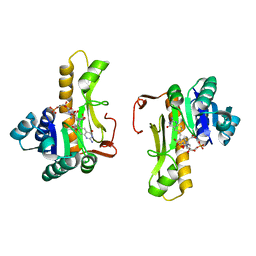 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLASE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-26 | | Release date: | 2000-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1U5L
 
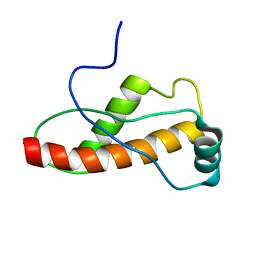 | | Solution Structure of the turtle prion protein fragment (121-226) | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-28 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chicken, turtle, and frog
PROC.NATL.ACAD.SCI.USA, 102, 2005
|
|
2FCG
 
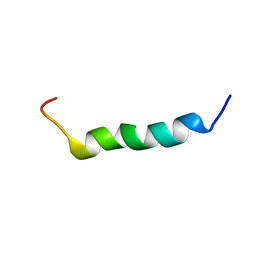 | | Solution structure of the C-terminal fragment of human LL-37 | | Descriptor: | Antibacterial protein FALL-39, core peptide | | Authors: | Wang, G, Li, X. | | Deposit date: | 2005-12-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Human LL-37 Fragments and NMR-Based Identification of a Minimal Membrane-Targeting Antimicrobial and Anticancer Region.
J.Am.Chem.Soc., 128, 2006
|
|
1SSV
 
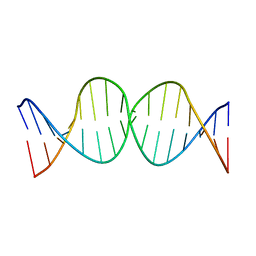 | | Compensating bends in a 16 base-pair DNA oligomer containing a T3A3 segment | | Descriptor: | 5'-D(*CP*GP*AP*GP*GP*TP*TP*TP*AP*AP*AP*CP*CP*TP*CP*G)-3' | | Authors: | McAteer, K, Aceves-Gaona, A, Michalczyk, R, Buchko, G.W, Isern, N.G, Silks, L.A, Miller, J.H, Kennedy, M.A. | | Deposit date: | 2004-03-24 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Compensating bends in a 16-base-pair DNA oligomer containing a T(3)A(3) segment: A NMR study of global DNA curvature
Biopolymers, 75, 2004
|
|
2JT3
 
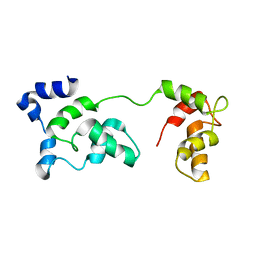 | | Solution Structure of F153W cardiac troponin C | | Descriptor: | Troponin C | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2JTZ
 
 | | Solution structure and chemical shift assignments of the F104-to-5-flurotryptophan mutant of cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
2FBU
 
 | | Solution structure of the N-terminal fragment of human LL-37 | | Descriptor: | Antibacterial protein FALL-39, core peptide | | Authors: | Wang, G, Li, X. | | Deposit date: | 2005-12-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Human LL-37 Fragments and NMR-Based Identification of a Minimal Membrane-Targeting Antimicrobial and Anticancer Region
J.Am.Chem.Soc., 128, 2006
|
|
2JT0
 
 | | Solution structure of F104W cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P.-J, Sykes, B.D. | | Deposit date: | 2007-07-17 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies.
Protein Sci., 14, 2005
|
|
2GZP
 
 | | Solution NMR structure of Q8ZP25 from Salmonella typhimurium LT2; Northeast Structural Genomics Consortium Target STR70 | | Descriptor: | putative chaperone | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T.B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-11 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Q8ZP24 from Salmonella typhimurium LT2.
To be Published
|
|
2JT8
 
 | | Solution structure of the F153-to-5-flurotryptophan mutant of human cardiac troponin C | | Descriptor: | Troponin C, slow skeletal and cardiac muscles | | Authors: | Wang, X, Mercier, P, Letourneau, P, Sykes, B.D. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Effects of Phe-to-Trp mutation and fluorotryptophan incorporation on the solution structure of cardiac troponin C, and analysis of its suitability as a potential probe for in situ NMR studies
Protein Sci., 14, 2005
|
|
1C3E
 
 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
2GB8
 
 | | Solution structure of the complex between yeast iso-1-cytochrome c and yeast cytochrome c peroxidase | | Descriptor: | Cytochrome c iso-1, Cytochrome c peroxidase, HEME C, ... | | Authors: | Volkov, A.N, Worrall, J.A.R, Ubbink, M. | | Deposit date: | 2006-03-10 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G57
 
 | | Structure of the Phosphorylation Motif of the oncogenic Protein beta-Catenin Recognized By a Selective Monoclonal Antibody | | Descriptor: | Beta-catenin | | Authors: | Megy, S, Bertho, G, Gharbi-Benarous, J, Baleux, F, Benarous, R, Girault, J.P. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | STD and TRNOESY NMR studies for the epitope mapping of the phosphorylation motif of the oncogenic protein beta-catenin recognized by a selective monoclonal antibody
Febs Lett., 580, 2006
|
|
2FGX
 
 | | Solution NMR Structure of Protein Ne2328 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeT3. | | Descriptor: | putative thioredoxin | | Authors: | Eletsky, A, Liu, G, Yee, A, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-22 | | Release date: | 2006-02-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Nitrosomonas Europaea Hypothetical Protein Ne2328: Northeast Structural Genomics Consortium Target NeT3
To be Published
|
|
2JRZ
 
 | | Solution structure of the Bright/ARID domain from the human JARID1C protein. | | Descriptor: | Histone demethylase JARID1C | | Authors: | Koehler, C, Bishop, S, Dowler, E.F, Diehl, A, Schmieder, P, Leidert, M, Sundstrom, M, Arrowsmith, C.H, Wiegelt, J, Edwards, A, Oschkinat, H, Ball, L.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein.
Biomol.Nmr Assign., 2, 2008
|
|
2KL3
 
 | | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A | | Descriptor: | Alr3790 protein | | Authors: | Eletsky, A, Belote, R.L, Ciccosanti, C, Janjua, H, Nair, R, Rost, B, Swapna, G.V.T, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the Rhodanese-like domain from Anabaena sp Alr3790 protein. Northeast Structural Genomics Consortium Target NsR437A
To be Published
|
|
2K5H
 
 | | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a | | Descriptor: | Conserved protein | | Authors: | Wu, Y, Singarapu, K, Semesi, A, Sukumaran, D, Yee, A, Garcia, M, Arrowsmith, C, Szyperski, T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by MTH693 from Methanobacterium thermoautotrophicum: Northeast Structural Genomics Consortium target tt824a
To be Published
|
|
