6U7U
 
 | | NMR solution structure of triazole bridged matriptase inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-THR-LYS-SER-ILE-PRO-PRO-ARG-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8DJ9
 
 | | Carbonic Anhydrase II in complex with Ibuprofen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, ... | | Authors: | Combs, J.C, McKenna, R, Andring, J.T. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Ibuprofen: a weak inhibitor of carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7S13
 
 | | Crystal structure of Fab in complex with mouse CD96 dimer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Lee, P.S, Barman, I, Strop, P. | | Deposit date: | 2021-08-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibody blockade of CD96 by distinct molecular mechanisms.
Mabs, 13, 2021
|
|
5KFZ
 
 | |
8WN6
 
 | |
7RW5
 
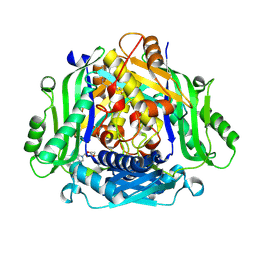 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
8AO3
 
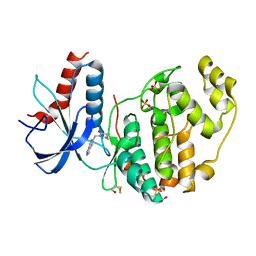 | | Specific covalent inhibitor of ERK2 | | Descriptor: | 2-chloranyl-~{N}-(1~{H}-indazol-5-ylmethyl)ethanamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.778 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8OXE
 
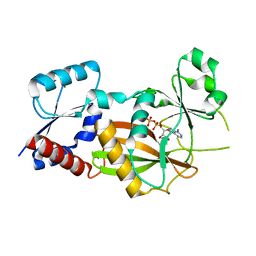 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
8WS7
 
 | |
9DUI
 
 | |
6VVJ
 
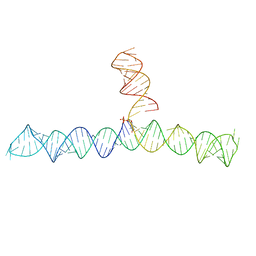 | | Cap1G-TPUA | | Descriptor: | RNA (130-MER) | | Authors: | Summers, M.F, Brown, J.D. | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for transcriptional start site control of HIV-1 RNA fate.
Science, 368, 2020
|
|
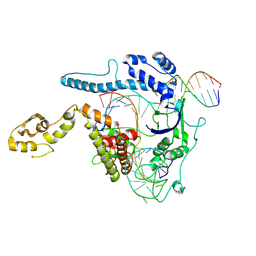
8WRR
 
 | |
9DHB
 
 | | STRUCTURE OF SERINE HYDROXYMETHYLTRANSFERASE 5 FROM GLYCINE MAX CULTIVAR ESSEX COMPLEXED WITH PLP-GLYCINE AND 5-FORMYLTETRAHYDROFOLATE | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Owuocha, L.F. | | Deposit date: | 2024-09-03 | | Release date: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | STRUCTURE OF SERINE HYDROXYMETHYLTRANSFERASE 5 FROM GLYCINE MAX CULTIVAR ESSEX COMPLEXED WITH PLP-GLYCINE AND 5-FORMYLTETRAHYDROFOLATE
To Be Published
|
|
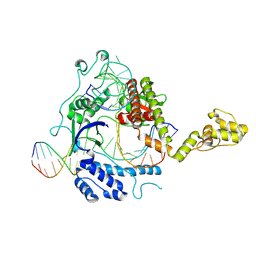
8WRQ
 
 | |
8WS9
 
 | |
8APF
 
 | | rotational state 2a of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
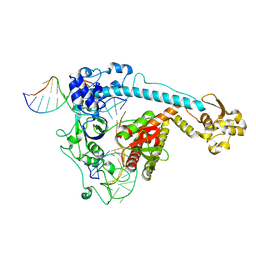
8WRS
 
 | |
8APD
 
 | | rotational state 1d of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8WRP
 
 | |
5XVU
 
 | | Crystal structure of the protein kinase CK2 catalytic domain from Plasmodium falciparum bound to ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | El Sahili, A, Lescar, J, Ruiz-Carrillo, D, Lin, J.Q. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The protein kinase CK2 catalytic domain from Plasmodium falciparum: crystal structure, tyrosine kinase activity and inhibition.
Sci Rep, 8, 2018
|
|
5KGI
 
 | |
9HRC
 
 | | Structure of MNK2 in complex with inhibitor NUCC-0201049 | | Descriptor: | 1-[5-chloranyl-4-[(3~{R})-pyrrolidin-3-yl]oxy-pyrimidin-2-yl]benzimidazole, GLYCEROL, MAP kinase-interacting serine/threonine-protein kinase 2, ... | | Authors: | Schiltz, G.E, Vagadia, P.V. | | Deposit date: | 2024-12-17 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.161 Å) | | Cite: | Discovery of Potent and Selective MNK Kinase Inhibitors for the Treatment of Leukemia.
J.Med.Chem., 68, 2025
|
|
6BG9
 
 | |
8AP6
 
 | | Trypanosoma brucei mitochondrial F1Fo ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
5GV1
 
 | | Crystal structure of ENZbleach xylanase wild type | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Chitnumsub, P, Jaruwat, A, Boonyapakorn, K, Noytanom, K. | | Deposit date: | 2016-09-01 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based protein engineering for thermostable and alkaliphilic enhancement of endo-beta-1,4-xylanase for applications in pulp bleaching
J. Biotechnol., 259, 2017
|
|
