7F8O
 
 | | Cryo-EM structure of the C-terminal deletion mutant of human PANX1 in a nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Pannexin-1 | | Authors: | Kuzuya, M, Hirano, H, Hayashida, K, Watanabe, M, Kobayashi, K, Tani, K, Fujiyoshi, Y, Oshima, A. | | Deposit date: | 2021-07-02 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human pannexin-1 in nanodiscs reveal gating mediated by dynamic movement of the N terminus and phospholipids.
Sci.Signal., 15, 2022
|
|
7FJ2
 
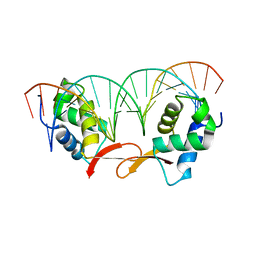 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
1UAJ
 
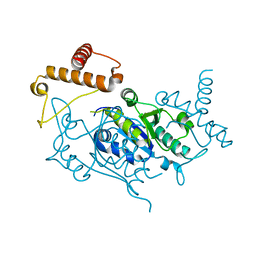 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UCD
 
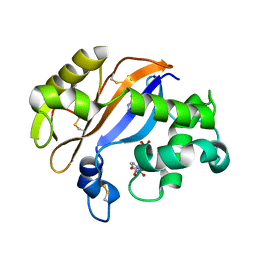 | | Crystal structure of Ribonuclease MC1 from bitter gourd seeds complexed with 5'-UMP | | Descriptor: | Ribonuclease MC, URACIL, URIDINE-5'-MONOPHOSPHATE | | Authors: | Suzuki, A, Numata, T, Yao, M, Kimura, M, Tanaka, I. | | Deposit date: | 2003-04-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of RNase MC1 from bitter gourd seeds in complex with 5'UMP
To be published
|
|
1UCQ
 
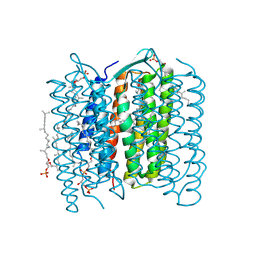 | | Crystal structure of the L intermediate of bacteriorhodopsin | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, RETINAL, ... | | Authors: | Kouyama, T, Nishikawa, T, Tokuhisa, T, Okumura, H. | | Deposit date: | 2003-04-17 | | Release date: | 2003-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the L Intermediate of Bacteriorhodopsin: Evidence for Vertical Translocation of a Water Molecule during the Proton Pumping Cycle.
J.Mol.Biol., 335, 2004
|
|
1UD3
 
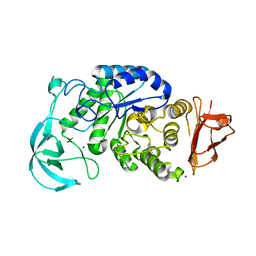 | | Crystal structure of AmyK38 N289H mutant | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
7F6V
 
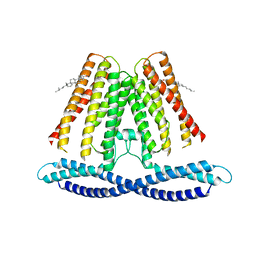 | | Cryo-EM structure of the human TACAN channel in a closed state | | Descriptor: | CHOLESTEROL, Ion channel TACAN | | Authors: | Chen, X.Z, Wang, Y.J, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2021-06-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structure of the human TACAN in a closed state.
Cell Rep, 38, 2022
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
1UGB
 
 | |
1UD8
 
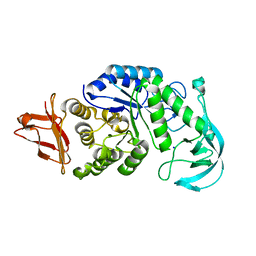 | | Crystal structure of AmyK38 with lithium ion | | Descriptor: | SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UDY
 
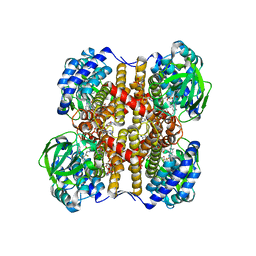 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
7FGF
 
 | |
7F3A
 
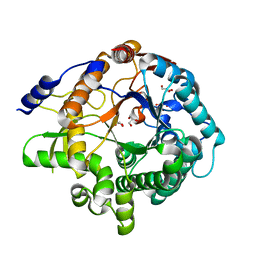 | | Arabidopsis thaliana GH1 beta-glucosidase AtBGlu42 | | Descriptor: | Beta-glucosidase 42, GLYCEROL | | Authors: | Horikoshi, S, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate specificity of glycoside hydrolase family 1 beta-glucosidase AtBGlu42 from Arabidopsis thaliana and its molecular mechanism.
Biosci.Biotechnol.Biochem., 86, 2022
|
|
7F7F
 
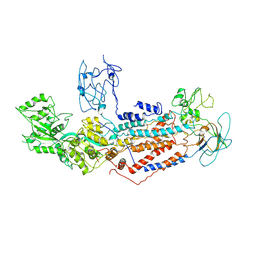 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
1U5I
 
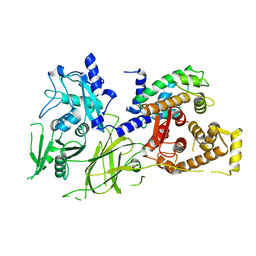 | | Crystal Structure analysis of rat m-calpain mutant Lys10 Thr | | Descriptor: | Calpain 2, large [catalytic] subunit precursor, Calpain small subunit 1 | | Authors: | Hosfield, C.M, Pal, G.P, Elce, J.S, Jia, Z. | | Deposit date: | 2004-07-27 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Activation of calpain by Ca2+: roles of the large subunit N-terminal and domain III-IV linker peptides
J.Mol.Biol., 343, 2004
|
|
7EPZ
 
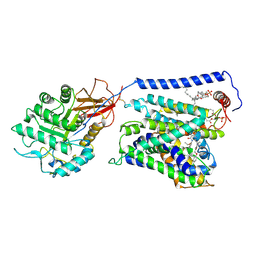 | | Overall structure of Erastin-bound xCT-4F2hc complex | | Descriptor: | 1,2-DISTEAROYL-SN-GLYCERO-3-PHOSPHATE, 2-[(1S)-1-[4-[2-(4-chloranylphenoxy)ethanoyl]piperazin-1-yl]ethyl]-3-(2-ethoxyphenyl)quinazolin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Li, Y.N, Zhang, Y.Y, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of erastin-bound xCT-4F2hc complex reveals molecular mechanisms underlying erastin-induced ferroptosis.
Cell Res., 32, 2022
|
|
1UAM
 
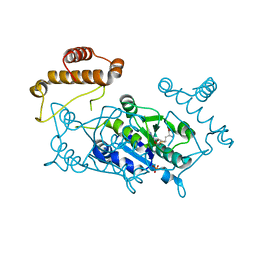 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
7F4V
 
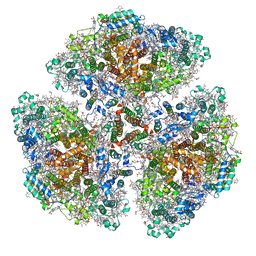 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
7F0I
 
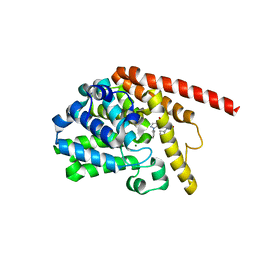 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
1UEH
 
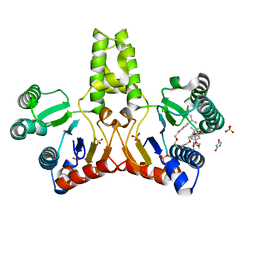 | | E. coli undecaprenyl pyrophosphate synthase in complex with Triton X-100, magnesium and sulfate | | Descriptor: | MAGNESIUM ION, OXTOXYNOL-10, SULFATE ION, ... | | Authors: | Chang, S.-Y, Ko, T.-P, Liang, P.-H, Wang, A.H.-J. | | Deposit date: | 2003-05-15 | | Release date: | 2003-08-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton
J.Biol.Chem., 278, 2003
|
|
7FG2
 
 | |
7FG3
 
 | |
7FG7
 
 | |
7F8Y
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with devazepide | | Descriptor: | N-[(3S)-1-methyl-2-oxidanylidene-5-phenyl-3H-1,4-benzodiazepin-3-yl]-1H-indole-2-carboxamide, fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
7F8U
 
 | | Crystal structure of the cholecystokinin receptor CCKAR in complex with lintitript | | Descriptor: | 2-[2-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]carbamoyl]indol-1-yl]ethanoic acid, Fusion protein of Cholecystokinin receptor type A and Endolysin | | Authors: | Zhang, X, He, C, Wang, M, Zhou, Q, Yang, D, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2021-07-02 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the human cholecystokinin receptors bound to agonists and antagonists.
Nat.Chem.Biol., 17, 2021
|
|
