3VQV
 
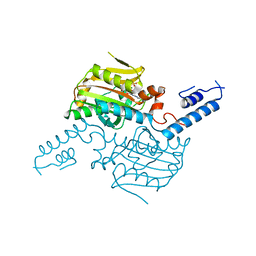 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with AMPPNP (re-refined) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3V8Y
 
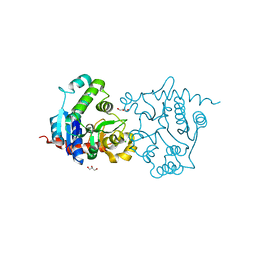 | | Structure of apo-glycogenin truncated at residue 270 | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1 | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3V91
 
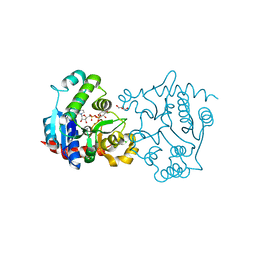 | | Structure of T82M glycogenin mutant truncated at residue 270 complexed with UDP-glucose | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycogenin-1, ... | | Authors: | Carrizo, M.E, Romero, J.M, Issoglio, F.M, Curtino, J.A. | | Deposit date: | 2011-12-23 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and biochemical insight into glycogenin inactivation by the glycogenosis-causing T82M mutation.
Febs Lett., 586, 2012
|
|
3WQE
 
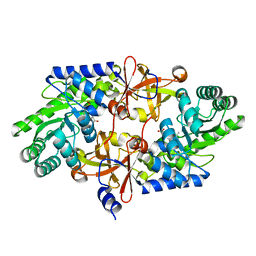 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-allothreonine | | Descriptor: | D-allothreonine, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WT4
 
 | | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa | | Descriptor: | CARBONATE ION, Probable M18 family aminopeptidase 2, ZINC ION | | Authors: | Nguyen, D.D, Pandian, R, Kim, D.D, Ha, S.C, Yoon, H.J, Kim, K.S, Yun, K.H, Kim, J.H, Kim, K.K. | | Deposit date: | 2014-04-07 | | Release date: | 2014-04-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic bases for the metal preference of the M18 aminopeptidase from Pseudomonas aeruginosa
Biochem.Biophys.Res.Commun., 447, 2014
|
|
3WQD
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 complexed with D-erythro-3-hydroxyaspartate | | Descriptor: | (3S)-3-hydroxy-D-aspartic acid, D-threo-3-hydroxyaspartate dehydratase, MAGNESIUM ION, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQF
 
 | |
3WQC
 
 | | D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23 | | Descriptor: | CHLORIDE ION, D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, ... | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3WQG
 
 | | D-threo-3-hydroxyaspartate dehydratase C353A mutant in the metal-free form | | Descriptor: | D-threo-3-hydroxyaspartate dehydratase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yasutake, Y, Matsumoto, Y, Wada, M. | | Deposit date: | 2014-01-25 | | Release date: | 2015-01-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate stereospecificity of D-threo-3-hydroxyaspartate dehydratase from Delftia sp. HT23: a useful enzyme for the synthesis of optically pure L-threo- and D-erythro-3-hydroxyaspartate
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3RJ6
 
 | |
3PJ7
 
 | |
3RLG
 
 | | Crystal structure of Loxosceles intermedia phospholipase D isoform 1 H12A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K. | | Deposit date: | 2011-04-19 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization and preliminary X-ray diffraction analysis of a class II phospholipase D from Loxosceles intermedia venom.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5U76
 
 | |
3RQC
 
 | | Crystal structure of the catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrott, N.L, Crennell, S.J, Hough, D.W, Danson, M.J, van den Elsen, J.M.H. | | Deposit date: | 2011-04-28 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.01 Å) | | Cite: | The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly.
Febs J., 279, 2012
|
|
3PJ5
 
 | |
5U70
 
 | |
3R44
 
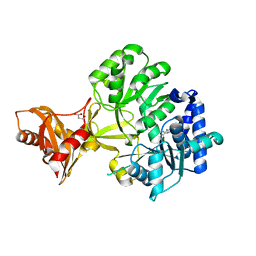 | | Mycobacterium tuberculosis fatty acyl CoA synthetase | | Descriptor: | HISTIDINE, MALONATE ION, fatty acyl CoA synthetase FADD13 (FATTY-ACYL-CoA SYNTHETASE) | | Authors: | Andersson, C.S, Martinez Molina, D, Hogbom, M. | | Deposit date: | 2011-03-17 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme.
Structure, 20, 2012
|
|
3SYU
 
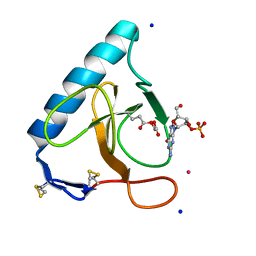 | | Re-refined coordinates for pdb entry 1det - ribonuclease T1 carboxymethylated at GLU 58 in complex with 2'GMP | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, Guanyl-specific ribonuclease T1, SODIUM ION, ... | | Authors: | Smart, O.S, Womack, T.O, Bricogne, G. | | Deposit date: | 2011-07-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3V56
 
 | | Re-refinement of PDB entry 1OSG - Complex between BAFF and a BR3 derived peptide presented in a beta-hairpin scaffold - reveals an additonal copy of the peptide. | | Descriptor: | BR3 derived peptive, SULFATE ION, Tumor necrosis factor ligand superfamily member 13B | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3URP
 
 | | Re-refinement of PDB entry 5RNT - ribonuclease T1 with guanosine-3',5'-diphosphate and phosphate ion bound | | Descriptor: | GUANOSINE-3',5'-DIPHOSPHATE, Guanyl-specific ribonuclease T1, PHOSPHATE ION, ... | | Authors: | Smart, O.S, Womack, T.O, Flensburg, C, Keller, P, Sharff, A, Paciorek, W, Vonrhein, C, Bricogne, G. | | Deposit date: | 2011-11-22 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4X3P
 
 | | Sirt2 in complex with a myristoyl peptide | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Wang, Y, Zhang, W, Hao, Q. | | Deposit date: | 2014-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Cell Chem Biol, 24, 2017
|
|
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
9MI7
 
 | | Crystal Structure of ADI-64597 ((human Fab, with substituted IgG1-CH1 (HC-L128R and K147R) and substituted kappa constant domain (LC-Q124E, V133Q, and T178R)) | | Descriptor: | ADI-64597 Fab heavy chain, ADI-64597 Fab light chain, CHLORIDE ION, ... | | Authors: | Battles, M.B, Welin, M, Laursen, M. | | Deposit date: | 2024-12-12 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Design of orthogonal constant domain interfaces to aid proper heavy/light chain pairing of bispecific antibodies.
Mabs, 17, 2025
|
|
7NPN
 
 | | B-brick bare in 5 mM Mg2+ | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bertosin, E, Stoemmer, P, Feigl, E, Wenig, M, Honemann, M, Dietz, H. | | Deposit date: | 2021-02-27 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10.38 Å) | | Cite: | Cryo-Electron Microscopy and Mass Analysis of Oligolysine-Coated DNA Nanostructures.
Acs Nano, 15, 2021
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
