1DUC
 
 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DV2
 
 | | The structure of biotin carboxylase, mutant E288K, complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BIOTIN CARBOXYLASE | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
1DRZ
 
 | | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | | Descriptor: | MAGNESIUM ION, PROTEIN (U1 SMALL RIBONUCLEOPROTEIN A), RNA (HEPATITIS DELTA VIRUS GENOMIC RIBOZYME), ... | | Authors: | Ferre-D'Amare, A.R, Zhou, K, Doudna, J.A. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a hepatitis delta virus ribozyme.
Nature, 395, 1998
|
|
1E0F
 
 | | Crystal structure of the human alpha-thrombin-haemadin complex: an exosite II-binding inhibitor | | Descriptor: | HAEMADIN, THROMBIN | | Authors: | Richardson, J.L, Kroeger, B, Hoefken, W, Pereira, P, Huber, R, Bode, W, Fuentes-Prior, P. | | Deposit date: | 2000-03-27 | | Release date: | 2000-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Human Alpha-Thrombin-Haemadin Complex: An Exosite II-Binding Inhibitor
Embo J., 19, 2000
|
|
1DWX
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1E1Q
 
 | | BOVINE MITOCHONDRIAL F1-ATPASE AT 100K | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, MAGNESIUM ION, ... | | Authors: | Braig, K, Menz, R.I, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2000-05-10 | | Release date: | 2000-06-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase Inhibited by Mg2+Adp and Aluminium Fluoride
Structure, 8, 2000
|
|
1E3U
 
 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
1DT5
 
 | |
1DUT
 
 | | FIV DUTP PYROPHOSPHATASE | | Descriptor: | DUTP PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Prasad, G.S, Stura, E.A, Mcree, D.E, Laco, G.S, Hasselkus-Light, C, Elder, J.H, Stout, C.D. | | Deposit date: | 1996-09-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus.
Protein Sci., 5, 1996
|
|
1DW9
 
 | | Structure of cyanase reveals that a novel dimeric and decameric arrangement of subunits is required for formation of the enzyme active site | | Descriptor: | CHLORIDE ION, CYANATE LYASE, SULFATE ION | | Authors: | Walsh, M.A, Otwinowski, Z, Perrakis, A, Anderson, P.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 1999-12-03 | | Release date: | 2000-05-16 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Cyanase Reveals that a Novel Dimeric and Decameric Arrangement of Subunits is Required for Formation of the Enzyme Active Site
Structure, 8, 2000
|
|
1DXL
 
 | | Dihydrolipoamide dehydrogenase of glycine decarboxylase from Pisum Sativum | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Faure, M, Cohen-Addad, C, Bourguignon, J, Macherel, D, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
1E2Y
 
 | |
1E0O
 
 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
1DUN
 
 | | EIAV DUTPASE NATIVE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DWV
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DVG
 
 | | CRYSTAL STRUCTURE OF RAT HEME OXYGENASE-1 IN COMPLEX WITH HEME; SELELENO-METHIONINE DERIVATIVE, MUTATED AT M51T,M93L,M155L,M191L. | | Descriptor: | HEME OXYGENASE-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Omata, Y, Kakuta, Y, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2000-01-20 | | Release date: | 2000-04-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of rat heme oxygenase-1 in complex with heme.
FEBS Lett., 471, 2000
|
|
1DQB
 
 | |
1DLO
 
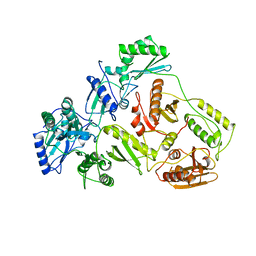 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Ding, J, Das, K, Hughes, S, Arnold, E. | | Deposit date: | 1996-04-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of unliganded HIV-1 reverse transcriptase at 2.7 A resolution: implications of conformational changes for polymerization and inhibition mechanisms.
Structure, 4, 1996
|
|
1DBV
 
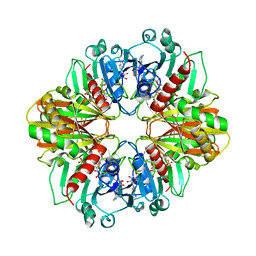 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
1DPG
 
 | |
1E2O
 
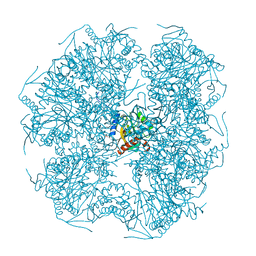 | | CATALYTIC DOMAIN FROM DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE | | Descriptor: | DIHYDROLIPOAMIDE SUCCINYLTRANSFERASE, SULFATE ION | | Authors: | Knapp, J.E, Mitchell, D.T, Yazdi, M.A, Ernst, S.R, Reed, L.J, Hackert, M.L. | | Deposit date: | 1998-05-26 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex.
J.Mol.Biol., 280, 1998
|
|
1DXY
 
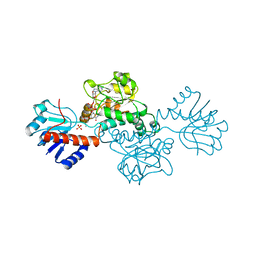 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
1DXX
 
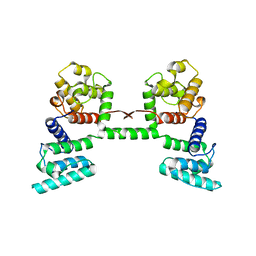 | |
1DYK
 
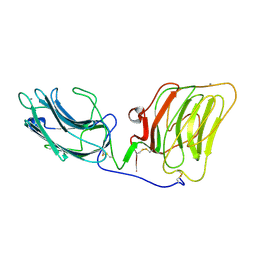 | | Laminin alpha 2 chain LG4-5 domain pair | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Tisi, D, Talts, J.F, Timple, R, Hohenester, E. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Laminin G-Like Domain Pair of the Laminin Alpha 2 Chain Harbouring Binding Sites for Alpha-Dystroglycan and Heparin
Embo J., 19, 2000
|
|
1E3K
 
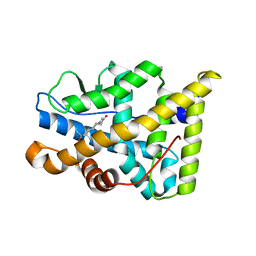 | | Human Progesteron Receptor Ligand Binding Domain in complex with the ligand metribolone (R1881) | | Descriptor: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, PROGESTERONE RECEPTOR | | Authors: | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Egner, U, Carrondo, M.A. | | Deposit date: | 2000-06-19 | | Release date: | 2001-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
