1E4K
 
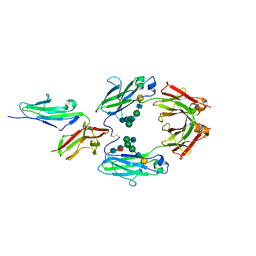 | | CRYSTAL STRUCTURE OF SOLUBLE HUMAN IGG1 FC FRAGMENT-FC-GAMMA RECEPTOR III COMPLEX | | Descriptor: | FC FRAGMENT OF HUMAN IGG1, LOW AFFINITY IMMUNOGLOBULIN GAMMA FC RECEPTOR III, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sondermann, P, Huber, R, Oosthuizen, V, Jacob, U. | | Deposit date: | 2000-07-07 | | Release date: | 2000-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The 3.2A Crystal Structure of the Human Igg1 Fc Fragment-Fc-Gamma-Riii Complex
Nature, 406, 2000
|
|
2EFG
 
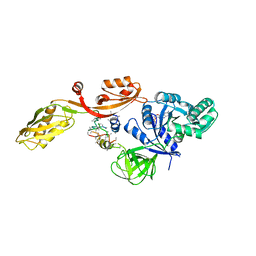 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1998-09-23 | | Release date: | 1999-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
2DYF
 
 | |
1RPO
 
 | |
1YDB
 
 | | STRUCTURAL BASIS OF INHIBITOR AFFINITY TO VARIANTS OF HUMAN CARBONIC ANHYDRASE II | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Nair, S.K, Christianson, D.W. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of inhibitor affinity to variants of human carbonic anhydrase II.
Biochemistry, 34, 1995
|
|
1RK2
 
 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP, SOLVED IN SPACE GROUP P212121 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Mowbray, S.L. | | Deposit date: | 1999-05-20 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Induced fit on sugar binding activates ribokinase.
J.Mol.Biol., 290, 1999
|
|
2EHT
 
 | |
1YDM
 
 | | X-Ray structure of Northeast Structural Genomics target SR44 | | Descriptor: | Hypothetical protein yqgN, SULFATE ION | | Authors: | Kuzin, A.P, Jianwei, S, Vorobiev, S, Acton, T, Xia, R, Ma, L.-C, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of Northeast Structural Genomics target SR44
To be Published
|
|
2EIA
 
 | |
2EIO
 
 | |
2DYT
 
 | |
1YJB
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YFM
 
 | |
1YF6
 
 | | Structure of a quintuple mutant of photosynthetic reaction center from rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Paddock, M.L, Chang, C, Xu, Q, Abresch, E.C, Axelrod, H.L. | | Deposit date: | 2004-12-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Quinone (Q(B)) Reduction by B-Branch Electron Transfer in Mutant Bacterial Reaction Centers from Rhodobacter sphaeroides: Quantum Efficiency and X-ray Structure.
Biochemistry, 44, 2005
|
|
2DZV
 
 | |
1YHA
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
2EPI
 
 | |
1Y6I
 
 | | Synechocystis GUN4 | | Descriptor: | Mg-chelatase cofactor GUN4 | | Authors: | Verdecia, M.A, Larkin, R.M, Ferrer, J.L, Riek, R, Chory, J, Noel, J.P. | | Deposit date: | 2004-12-06 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding
Plos Biol., 3, 2005
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2DZT
 
 | |
1V6C
 
 | | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11 | | Descriptor: | CALCIUM ION, SULFATE ION, alkaline serine protease, ... | | Authors: | Dong, D, Ihara, T, Motoshima, H, Watanabe, K. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11
To be Published
|
|
1V9I
 
 | |
1YB7
 
 | | Hydroxynitrile lyase from hevea brasiliensis in complex with 2,3-dimethyl-2-hydroxy-butyronitrile | | Descriptor: | (S)-2-HYDROXY-2,3-DIMETHYLBUTANENITRILE, (S)-acetone-cyanohydrin lyase, SULFATE ION | | Authors: | Gruber, K, Gartler, G, Kratky, C. | | Deposit date: | 2004-12-20 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis
J.Biotechnol., 129, 2007
|
|
2EET
 
 | | Guanine Riboswitch A21G, U75C mutant bound to hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine Riboswitch, ... | | Authors: | Gilbert, S.D, Edwards, A.L, Batey, R.T. | | Deposit date: | 2007-02-18 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational analysis of the purine riboswitch aptamer domain
Biochemistry, 46, 2007
|
|
1GAV
 
 | | BACTERIOPHAGE GA PROTEIN CAPSID | | Descriptor: | BACTERIOPHAGE GA PROTEIN CAPSID | | Authors: | Tars, K, Bundule, M, Liljas, L. | | Deposit date: | 1997-01-28 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses.
J.Mol.Biol., 271, 1997
|
|
