1S7O
 
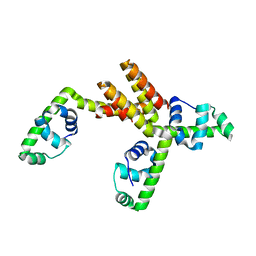 | | Crystal structure of putative DNA binding protein SP_1288 from Streptococcus pygenes | | Descriptor: | Hypothetical UPF0122 protein SPy1201/SpyM3_0842/SPs1042/spyM18_1152 | | Authors: | Oganesyan, V, Pufan, R, DeGiovanni, A, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-01-29 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the putative DNA-binding protein SP_1288 from Streptococcus pyogenes.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S7P
 
 | | Solution structure of thermolysin digested microcin J25 | | Descriptor: | microcin J25 | | Authors: | Rosengren, K.J, Blond, A, Afonso, C, Tabet, J.C, Rebuffat, S, Craik, D.J. | | Deposit date: | 2004-01-30 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Structure of thermolysin cleaved microcin J25: extreme stability of a two-chain antimicrobial peptide devoid of covalent links
Biochemistry, 43, 2004
|
|
1S7Q
 
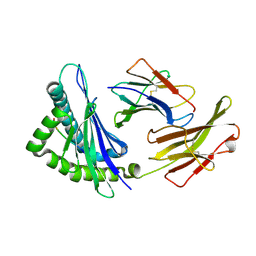 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7R
 
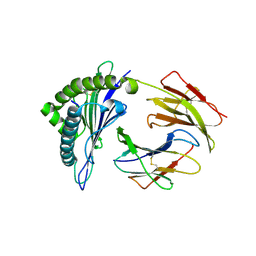 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7S
 
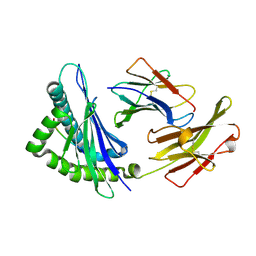 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7T
 
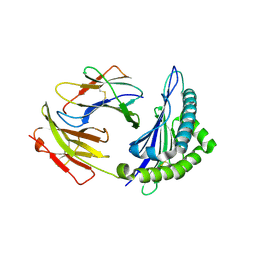 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7U
 
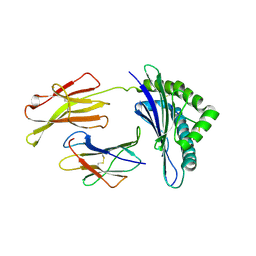 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7V
 
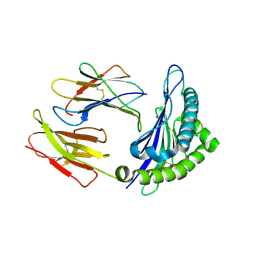 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7W
 
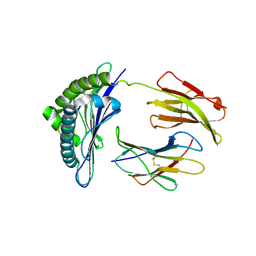 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7X
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1S7Y
 
 | |
1S7Z
 
 | | Structure of Ocr from Bacteriophage T7 | | Descriptor: | CESIUM ION, Gene 0.3 protein | | Authors: | Walkinshaw, M.D, Taylor, P, Sturrock, S.S, Atanasiu, C, Berg, T, Henderson, R.M, Edwardson, J.M, Dryden, D.T. | | Deposit date: | 2004-01-30 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of Ocr from Bacteriophage T7, a Protein that Mimics B-Form DNA
Mol.Cell, 9, 2002
|
|
1S80
 
 | | Structure of Serine Acetyltransferase from Haemophilis influenzae Rd | | Descriptor: | Serine acetyltransferase | | Authors: | Gorman, J, Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-01-30 | | Release date: | 2004-08-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of serine acetyltransferase from Haemophilus influenzae Rd.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S81
 
 | | PORCINE TRYPSIN WITH NO INHIBITOR BOUND | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S82
 
 | | PORCINE TRYPSIN COMPLEXED WITH BORATE AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, 1,3,2-DIOXABOROLAN-2-OL, CALCIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S83
 
 | | PORCINE TRYPSIN COMPLEXED WITH 4-AMINO PROPANOL | | Descriptor: | 4-HYDROXYBUTAN-1-AMINIUM, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, DeRose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S84
 
 | | PORCINE TRYPSIN COVALENT COMPLEX WITH 4-AMINO BUTANOL, BORATE AND ETHYLENE GLYCOL | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3,2-DIOXABOROLAN-2-YLOXY)BUTAN-1-AMINIUM, CALCIUM ION, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S85
 
 | | PORCINE TRYPSIN COMPLEXED WITH P-HYDROXYMETHYL BENZAMIDINE AND BORATE | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Transue, T.R, Krahn, J.M, Gabel, S.A, Derose, E.F, London, R.E. | | Deposit date: | 2004-01-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Biochemistry, 43, 2004
|
|
1S88
 
 | | NMR structure of a DNA duplex with two INA nucleotides inserted opposite each other, dCTCAACXCAAGCT:dAGCTTGXGTTGAG | | Descriptor: | 5'-D(*AP*GP*CP*TP*TP*GP*(2DM)P*GP*TP*TP*GP*AP*G)-3', 5'-D(*CP*TP*CP*AP*AP*CP*(2DM)P*CP*AP*AP*GP*CP*T)-3' | | Authors: | Nielsen, C.B, Petersen, M, Pedersen, E.B, Hansen, P.E, Christensen, U.B. | | Deposit date: | 2004-01-31 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a modified DNA oligonucleotide containing a new intercalating nucleic acid.
Bioconjug.Chem., 15, 2004
|
|
1S89
 
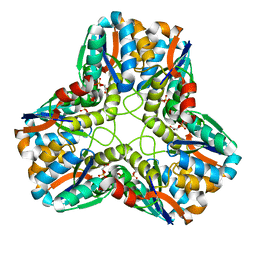 | |
1S8A
 
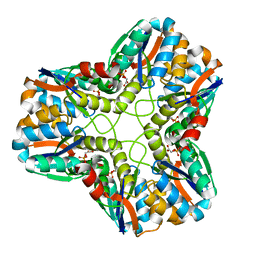 | |
1S8C
 
 | | Crystal structure of human heme oxygenase in a complex with biliverdine | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1 | | Authors: | Lad, L, Friedman, J, Li, H, Bhaskar, B, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Human Heme Oxygenase-1 in a Complex with Biliverdin
Biochemistry, 43, 2004
|
|
1S8D
 
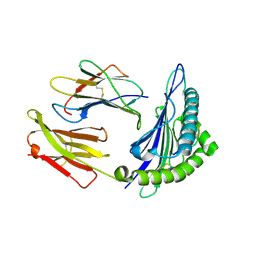 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9-3A | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-02-02 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1S8E
 
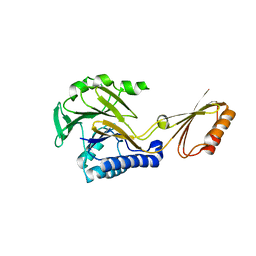 | | Crystal structure of Mre11-3 | | Descriptor: | MANGANESE (II) ION, exonuclease putative | | Authors: | Hopfner, K.P. | | Deposit date: | 2004-02-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of Mre11-3
Nucleic Acids Res., 32, 2004
|
|
1S8F
 
 | |
