4QGB
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 | | Descriptor: | 50S ribosomal protein L1, ACETATE ION, CHLORIDE ION | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5YDU
 
 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
6MD3
 
 | | Structure of T. brucei RRP44 PIN domain | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Rrp44p homologue, ... | | Authors: | Guimaraes, B.G, Cesaro, G. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Trypanosoma brucei RRP44 is involved in an early stage of large ribosomal subunit RNA maturation.
RNA Biol, 16, 2019
|
|
5LTA
 
 | |
6QWL
 
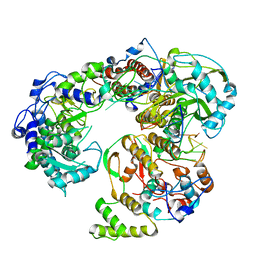 | | Influenza B virus (B/Panama/45) polymerase Hetermotrimer in complex with 3'5' cRNA promoter | | Descriptor: | 3' cRNA, 5' cRNA, Polymerase acidic protein, ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
6RR7
 
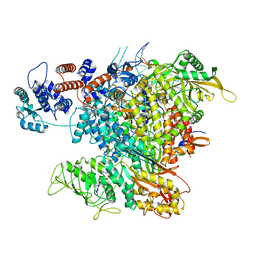 | | Influenza A virus (A/NT/60/1968) polymerase Heterotrimer bound to 3'5' vRNA promoter and capped RNA primer | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(M7G))-R(P*AP*AP*UP*CP*U)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-05-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
8OST
 
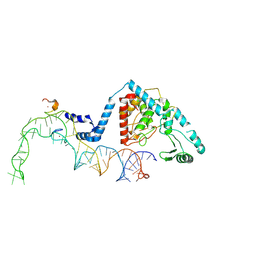 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6QX3
 
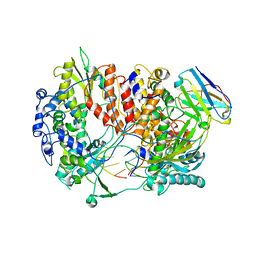 | | Influenza A virus (A/NT/60/1968) polymerase Hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
3EPL
 
 | |
3EPJ
 
 | |
3EPK
 
 | |
3EPH
 
 | |
6QX8
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
9C7A
 
 | | Crystal structure of R149W neurodevelopmental disease-associated U2AF2 variant | | Descriptor: | DNA/RNA (5'-R(P*UP*U)-D(P*U)-R(P*UP*U)-D(P*(BRU))-R(P*CP*C)-3'), GLYCEROL, SODIUM ION, ... | | Authors: | Maji, D, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2024-06-10 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recurrent Neurodevelopmentally Associated Variants of the Pre-mRNA Splicing Factor U2AF2 Alter RNA Binding Affinities and Interactions.
Biochemistry, 63, 2024
|
|
9C7B
 
 | |
6QXE
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
1VQ7
 
 | |
1VQN
 
 | |
1VQ6
 
 | |
4WYK
 
 | |
8UAU
 
 | | human ATE1 in complex with Arg-tRNA and a peptide substrate | | Descriptor: | Isoform ATE1-2 of Arginyl-tRNA--protein transferase 1, RNA (76-MER), ZINC ION, ... | | Authors: | Huang, W, Zhang, Y, Taylor, D.J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Oligomerization and a distinct tRNA-binding loop are important regulators of human arginyl-transferase function.
Nat Commun, 15, 2024
|
|
7NK1
 
 | | 1918 Influenza virus polymerase heterotirmer in complex with vRNA promoters and Nb8201 | | Descriptor: | Nanobody8201, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
5DTO
 
 | | Dengue virus full length NS5 complexed with viral Cap 0-RNA and SAH | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Zhao, Y, Soh, T.S, Lim, S.P, Chung, K.Y, Swaminathan, K, Vasudevan, S.G, Shi, P.-Y, Lescar, J, Luo, D. | | Deposit date: | 2015-09-18 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5)
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5M52
 
 | | Crystal structure of yeast Brr2 full-lenght in complex with Prp8 Jab1 domain | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | Authors: | Wollenhaupt, J, Absmeier, E, Becke, C, Santos, K.F, Wahl, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2.
Cell Cycle, 16, 2017
|
|
7NYB
 
 | | TrmB complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
