8HCJ
 
 | | Structure of GH43 family enzyme, Xylan 1, 4 Beta- xylosidase from pseudopedobacter saltans | | Descriptor: | CALCIUM ION, Xylan 1,4-beta-xylosidase | | Authors: | Vishwakarma, P, Sachdeva, E, Goyal, A, Ethayathulla, A.S, Das, U, Kaur, P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.566 Å) | | Cite: | Deciphering the structural and biochemical aspects of xylosidase from Pseudopedobacter saltans.
Int.J.Biol.Macromol., 291, 2024
|
|
2NAO
 
 | | Atomic resolution structure of a disease-relevant Abeta(1-42) amyloid fibril | | Descriptor: | Beta-amyloid protein 42 | | Authors: | Waelti, M.A, Ravotti, F, Arai, H, Glabe, C, Wall, J, Bockmann, A, Guntert, P, Meier, B.H, Riek, R. | | Deposit date: | 2016-01-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Atomic-resolution structure of a disease-relevant A beta (1-42) amyloid fibril.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6NEB
 
 | | MYC Promoter G-Quadruplex with 1:6:1 loop length | | Descriptor: | DNA (27-MER) | | Authors: | Dickerhoff, J, Onel, B, Chen, L, Chen, Y, Yang, D. | | Deposit date: | 2018-12-17 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length.
Acs Omega, 4, 2019
|
|
1TTB
 
 | |
9CLQ
 
 | | PANK3 complex structure with compound PZ-4352 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-cyclopropyl-2-fluorophenyl)-1-[4-(6-fluoro[1,3]oxazolo[4,5-b]pyridin-2-yl)piperazin-1-yl]ethan-1-one, ACETATE ION, ... | | Authors: | Yun, M, Lee, R.E. | | Deposit date: | 2024-07-12 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PANK3 complex structure with compound PZ-4352
To Be Published
|
|
4Y3Z
 
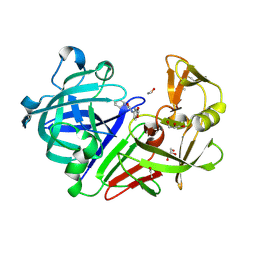 | | Endothiapepsin in complex with fragment 41 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
9CT7
 
 | | Tricomplex of Compound 1, KRAS G12D, and CypA | | Descriptor: | (2R)-2-cyclopentyl-N-[(1M,8S,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]-2-(N-methylacetamido)acetamide (non-preferred name), CHLORIDE ION, Isoform 2B of GTPase KRas, ... | | Authors: | Zhang, D, Bar Ziv, T, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
4Y56
 
 | | Endothiapepsin in complex with fragment 58 | | Descriptor: | 1,2-ETHANEDIOL, 1-(1,3-dimethyl-1H-pyrazol-5-yl)methanamine, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4EW2
 
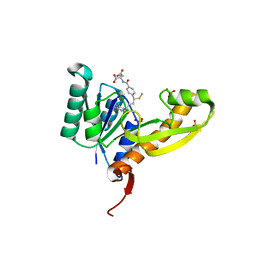 | | The structure of human glycinamide ribonucleotide transformylase in complex with 10S-methylthio-DDATHF. | | Descriptor: | N-({4-[(1S)-4-(2,4-diamino-6-oxo-1,6-dihydropyrimidin-5-yl)-1-(methylsulfanyl)butyl]phenyl}carbonyl)-L-glutamic acid, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Connelly, S, DeMartino, K, Boger, D.L, Wilson, I.A. | | Deposit date: | 2012-04-26 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Biological and Structural Evaluation of 10R- and 10S-Methylthio-DDACTHF Reveals a New Role for Sulfur in Inhibition of Glycinamide Ribonucleotide Transformylase.
Biochemistry, 52, 2013
|
|
2NAW
 
 | |
4E77
 
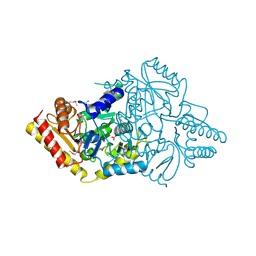 | | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92 | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase, NITRATE ION, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, W, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
4E8D
 
 | | Crystal structure of streptococcal beta-galactosidase | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35 | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
8HSA
 
 | |
7WJO
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
8HUB
 
 | | AMP deaminase 2 in complex with an inhibitor | | Descriptor: | 3,3-dimethyl-4-(phenylmethyl)-2~{H}-quinoxaline-1-carboxamide, AMP deaminase 2, ZINC ION | | Authors: | Adachi, T, Doi, S. | | Deposit date: | 2022-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The discovery of 3,3-dimethyl-1,2,3,4-tetrahydroquinoxaline-1-carboxamides as AMPD2 inhibitors with a novel mechanism of action.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
4Y1Y
 
 | | Complex of human Galectin-1 and (6OSO3)Galbeta1-3GlcNAc | | Descriptor: | Galectin-1, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranoside | | Authors: | Lin, H.Y, Hsieh, T.J, Lin, C.H. | | Deposit date: | 2015-02-09 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis of human galectin-1 inhibition with Ki values in the micro- to nanomolar range
To Be Published
|
|
6NRI
 
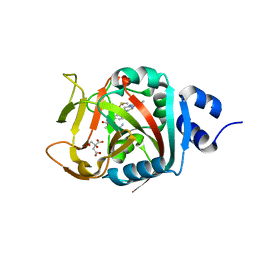 | | Crystal Structure of human PARP-1 ART domain bound to inhibitor UTT83 | | Descriptor: | (2Z)-2-{[4-(3-cyclopropyl-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carbonyl)phenyl]methylidene}-3-oxo-2,3-dihydro-1-benzofuran-7-carboxamide, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
9CRC
 
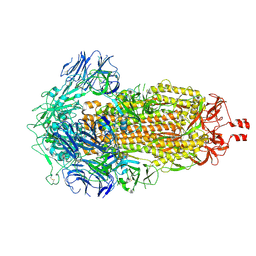 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRH
 
 | |
9CRI
 
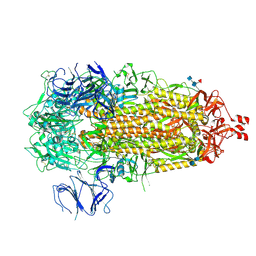 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Mu (B.1.621) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRD
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: B.1 variant 1 open RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRE
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Alpha (B.1.1.7) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Croll, T.I, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
9CRG
 
 | | Cryo-EM structure of SARS-CoV-2 Spike Proteins on intact virions: Gamma (P.1) variant 3 closed RBDs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ke, Z, Kotecha, A, Briggs, J.A.G. | | Deposit date: | 2024-07-22 | | Release date: | 2024-11-27 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Virion morphology and on-virus spike protein structures of diverse SARS-CoV-2 variants.
Embo J., 43, 2024
|
|
6NEA
 
 | | Human Acetylcholinesterase in complex with reactivator, HLo7 | | Descriptor: | 1-[({2,4-BIS[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]-4-CARBAMOYLPYRIDINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bester, S.M, McGuire, J, Height, J.J, Pegan, S.D. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.419 Å) | | Cite: | Synthesis and Molecular Properties of Nerve Agent Reactivator HLo-7 Dimethanesulfonate.
Acs Med.Chem.Lett., 10, 2019
|
|
2MRP
 
 | |
