2Z70
 
 | | E.coli RNase 1 in complex with d(CGCGATCGCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DTP*DCP*DGP*DCP*DG)-3'), Ribonuclease I | | Authors: | Martinez-Rodriguez, S, Loris, R, Messens, J. | | Deposit date: | 2007-08-09 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli
Protein Sci., 17, 2008
|
|
2PYB
 
 | |
2EKE
 
 | |
2FJC
 
 | | Crystal structure of antigen TpF1 from Treponema pallidum | | Descriptor: | Antigen TpF1, FE (III) ION | | Authors: | Thumiger, A, Polenghi, A, Papinutto, E, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of antigen TpF1 from Treponema pallidum.
Proteins, 62, 2006
|
|
1UE2
 
 | | Crystal structure of d(GC38GAAAGCT) | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*AP*AP*GP*CP*T)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanaba, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UB8
 
 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-03-31 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3RPG
 
 | | Bmi1/Ring1b-UbcH5c complex structure | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Bentley, M.L, Dong, K.C, Cochran, A.G. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6485 Å) | | Cite: | Recognition of UbcH5c and the nucleosome by the Bmi1/Ring1b ubiquitin ligase complex.
Embo J., 30, 2011
|
|
6HDE
 
 | | Structure of Escherichia coli dUTPase Q93H mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Benedek, A, Vertessy, B.G, Leveles, I. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Role of a Key Amino Acid Position in Species-Specific Proteinaceous dUTPase Inhibition.
Biomolecules, 9, 2019
|
|
1UE4
 
 | | Crystal structure of d(GCGAAAGC) | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ZZK
 
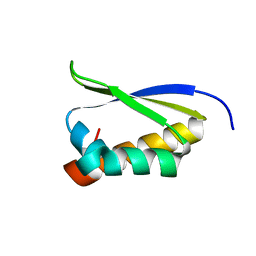 | | Crystal Structure of the third KH domain of hnRNP K at 0.95A resolution | | Descriptor: | Heterogeneous nuclear ribonucleoprotein K | | Authors: | Backe, P.H, Messias, A.C, Ravelli, R.B, Sattler, M, Cusack, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | X-Ray Crystallographic and NMR Studies of the Third KH Domain of hnRNP K in Complex with Single-Stranded Nucleic Acids
STRUCTURE, 13, 2005
|
|
1U01
 
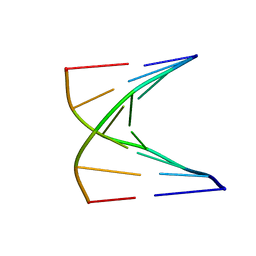 | | High resolution NMR structure of 5-d(GCGT*GCG)-3/5-d(CGCACGC)-3 (T*represents a cyclohexenyl nucleotide) | | Descriptor: | 5'-D(*CP*GP*CP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*(XTR)P*GP*CP*G)-3' | | Authors: | Nauwelaerts, K, Lescrinier, E, Sclep, G, Herdewijn, P. | | Deposit date: | 2004-07-12 | | Release date: | 2005-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cyclohexenyl nucleic acids: conformationally flexible oligonucleotides.
Nucleic Acids Res., 33, 2005
|
|
1UE3
 
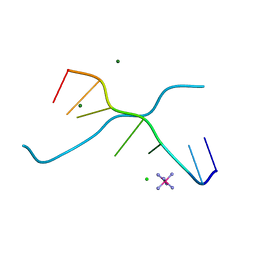 | | Crystal structure of d(GCGAAAGC) containing hexaamminecobalt | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
7BGS
 
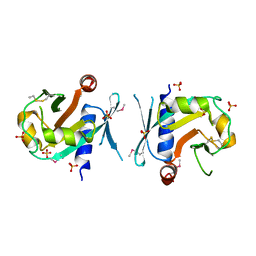 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-08 | | Release date: | 2022-01-19 | | Last modified: | 2022-02-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7BNX
 
 | | Archeal holliday junction resolvase from Thermus thermophilus phage 15-6 | | Descriptor: | Holliday junction resolvase, SULFATE ION | | Authors: | Hakansson, M, Ahlqvist, J, Linares Pasten, J.A, Jasilionis, A, Nordberg Karlsson, E, Al-Karadaghi, S. | | Deposit date: | 2021-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Crystal structure and initial characterization of a novel archaeal-like Holliday junction-resolving enzyme from Thermus thermophilus phage Tth15-6.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1WAO
 
 | | PP5 structure | | Descriptor: | MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Barford, D. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Tpr Domain-Mediated Regulation of Protein Phosphatase 5
Embo J., 24, 2005
|
|
1Y0X
 
 | | Thyroxine-Thyroid Hormone Receptor Interactions | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, CACODYLATE ION, Thyroid hormone receptor beta-1 | | Authors: | Sandler, B, Webb, P, Apriletti, J.W, Huber, B.R, Togashi, M, Cunha Lima, S.T, Juric, S, Nilsson, S, Wagner, R, Fletterick, R.J, Baxter, J.D. | | Deposit date: | 2004-11-16 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thyroxine-thyroid hormone receptor interactions.
J.Biol.Chem., 279, 2004
|
|
1XZX
 
 | | Thyroxine-Thyroid Hormone Receptor Interactions | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, CACODYLATE ION, Thyroid hormone receptor beta-1 | | Authors: | Sandler, B, Webb, P, Apriletti, J.W, Huber, B.R, Togashi, M, Cunha Lima, S.T, Juric, S, Nilsson, S, Wagner, R, Fletterick, R.J, Baxter, J.D. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Thyroxine-thyroid hormone receptor interactions.
J.Biol.Chem., 279, 2004
|
|
1Z60
 
 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | Descriptor: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | Authors: | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
3KF8
 
 | | Crystal structure of C. tropicalis Stn1-ten1 complex | | Descriptor: | Protein stn1, Protein ten1 | | Authors: | Sun, J, Yu, E.Y, Yang, Y.T, Confer, L.A, Sun, S.H, Wan, K, Lue, N.F, Lei, M. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres.
Genes Dev., 23, 2009
|
|
5WI8
 
 | |
5WIW
 
 | |
3OAH
 
 | |
1YVU
 
 | | Crystal structure of A. aeolicus Argonaute | | Descriptor: | CALCIUM ION, hypothetical protein aq_1447 | | Authors: | Yuan, Y.R, Pei, Y, Ma, J.B, Kuryavyi, V, Zhadina, M, Meister, G, Chen, H.Y, Dauter, Z, Tuschl, T, Patel, D.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of A. aeolicus Argonaute provides unique perspectives into the mechanism of guide strand-mediated mRNA cleavage
Mol.Cell, 19, 2005
|
|
2ACH
 
 | | CRYSTAL STRUCTURE OF CLEAVED HUMAN ALPHA1-ANTICHYMOTRYPSIN AT 2.7 ANGSTROMS RESOLUTION AND ITS COMPARISON WITH OTHER SERPINS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA 1-ANTICHYMOTRYPSIN, PHOSPHATE ION, ... | | Authors: | Baumann, U, Huber, R, Bode, W, Grosse, D, Lesjak, M, Laurell, C.B. | | Deposit date: | 1993-04-26 | | Release date: | 1993-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of cleaved human alpha 1-antichymotrypsin at 2.7 A resolution and its comparison with other serpins.
J.Mol.Biol., 218, 1991
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
