7FUG
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 5-benzyl-2-[[2-chloro-5-(1-methylpyrazol-3-yl)phenyl]methylamino]-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-5-benzyl-2-({[(5P)-2-chloro-5-(1-methyl-1H-pyrazol-3-yl)phenyl]methyl}amino)[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUH
 
 | |
7FUI
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with (Z)-2-cyano-N-[4-(3-fluorophenyl)-5-methylsulfonylpyrimidin-2-yl]-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide | | Descriptor: | (2Z)-2-cyano-N-[(4P)-4-(3-fluorophenyl)-5-(methanesulfonyl)pyrimidin-2-yl]-3-hydroxy-3-(5-methyl-1,2-oxazol-4-yl)prop-2-enamide, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUK
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(2-chloro-4-methylphenyl)-3-(2-phenylethyl)benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-5-(2-chloro-4-methylphenyl)-1-(2-phenylethyl)-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUL
 
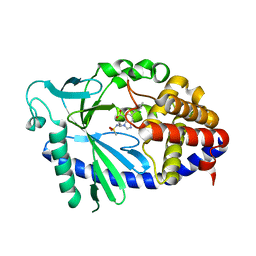 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 6-(2-chloro-4-methylphenyl)-3-[3-(methanesulfonamido)propyl]benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-5-(2-chloro-4-methylphenyl)-1-{3-[(methanesulfonyl)amino]propyl}-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Galley, G, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUM
 
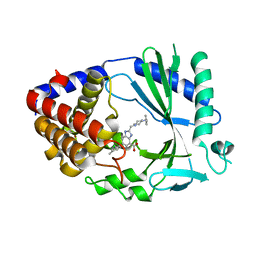 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(4-benzylpiperazin-1-yl)methyl]-6-(2-chloro-4-methylphenyl)-1H-benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-2-[(4-benzylpiperazin-1-yl)methyl]-5-(2-chloro-4-methylphenyl)-1H-benzimidazole-7-carboxylic acid, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Galley, G, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUN
 
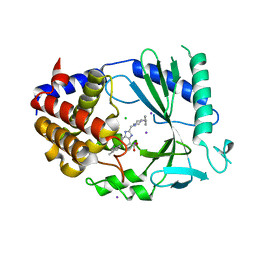 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(4-benzylpiperazin-1-yl)methyl]-6-(2-chloro-4-methylphenyl)-1H-benzimidazole-4-carboxylic acid | | Descriptor: | (5M)-2-[(4-benzylpiperazin-1-yl)methyl]-5-(2-chloro-4-methylphenyl)-1H-benzimidazole-7-carboxylic acid, CHLORIDE ION, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Galley, G, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7FUO
 
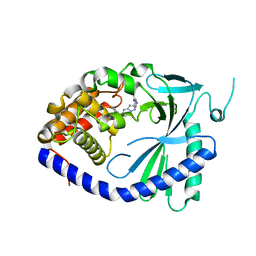 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(2-chlorophenyl)methylamino]-5-[(2-fluoroanilino)methyl]-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8R)-2-{[(2-chlorophenyl)methyl]amino}-5-[(2-fluoroanilino)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
7JXA
 
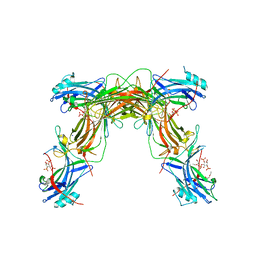 | | CRYSTAL STRUCTURE OF NATIVE BOVINE ARRESTIN 1 IN COMPLEX WITH INOSITOL 1,4,5-TRIPHOSPHATE | | Descriptor: | 2-ETHOXYETHANOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, S-arrestin, ... | | Authors: | Sander, C.L, Palczewski, K, Kiser, P.D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
7JIR
 
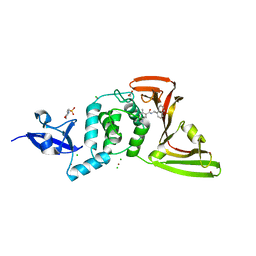 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIT
 
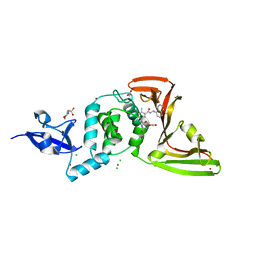 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder495 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(carbamoylcarbamoyl)amino]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JIW
 
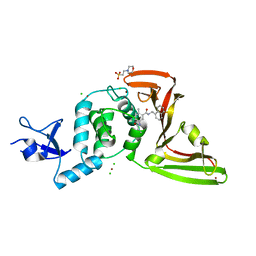 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
