4UMG
 
 | | Crystal structure of the Lin-41 filamin domain | | Descriptor: | PROTEIN LIN-41 | | Authors: | Tocchini, C, Keusch, J.J, Miller, S.B, Finger, S, Gut, H, Stadler, M, Ciosk, R. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Trim-Nhl Protein Lin-41 Controls the Onset of Developmental Plasticity in Caenorhabditis Elegans.
Plos Genet., 10, 2014
|
|
4UFZ
 
 | | Synthesis of Novel NAD Dependant DNA Ligase Inhibitors via Negishi Cross-Coupling: Development of SAR and Resistance Studies | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 5,7-bis(azanyl)-2-tert-butyl-4-(1,3-thiazol-2-yl)pyrido[2,3-d]pyrimidine-6-carbonitrile, DNA LIGASE | | Authors: | Murphy-Benenato, K.E, Boriack-Sjodin, P.A, Martinez-Botella, G, Carcanague, D, Gingipali, L, Gowravaram, M, Harang, J, Hale, M, Ioannidis, G, Jahic, H, Johnstone, M, Kutschke, A, Laganas, V.A, Loch, J, Oguto, H, Patel, S.J. | | Deposit date: | 2015-03-20 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Negishi Cross-Coupling Enabled Synthesis of Novel Nad(+)-Dependent DNA Ligase Inhibitors and Sar Development.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4UMP
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 3-(isoquinolin-7-yl)prop-2-yn-1-ol, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UI3
 
 | | Crystal structure of human tankyrase 2 in complex with TA-26 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-3H-quinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UHT
 
 | |
4UMR
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 4-fluoro-N-(1,2,3,4-tetrahydroisoquinolin-7-yl)benzamide, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
4UI5
 
 | | Crystal structure of human tankyrase 2 in complex with TA-41 | | Descriptor: | 8-methoxy-2-(4-methylphenyl)-3,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
7O77
 
 | | Structure of the PL6 family alginate lyase Patl3640 from Pseudoalteromonas atlantica T6c | | Descriptor: | GLYCEROL, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
4UHG
 
 | | Crystal structure of human tankyrase 2 in complex with TA-21 | | Descriptor: | 2-(4-METHYLPHENYL)-3,4-DIHYDROQUINAZOLIN-4-ONE, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Activity Relationships of 2-Arylquinazolin-4-Ones as Highly Selective and Potent Inhibitors of the Tankyrases.
Eur.J.Med.Chem., 118, 2016
|
|
4UIZ
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 7-(3,4-dimethoxyphenyl)-2-(4-methanesulfonylpiperazine-1-carbonyl)-5-methyl-4H,5H-thieno-3,2-c- pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,4-dimethoxyphenyl)-5-methyl-2-(4-methylsulfonylpiperazin-1-yl)carbonyl-thieno[3,2-c]pyridin-4-one, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C, Theodoulou, N.T, Bamborough, P, Humphreys, P.G. | | Deposit date: | 2015-04-03 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | The Discovery of I-Brd9, a Selective Cell Active Chemical Probe for Bromodomain Containing Protein 9 Inhibition.
J.Med.Chem., 59, 2016
|
|
4UR3
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans P2(1) crystal form | | Descriptor: | IRON/SULFUR CLUSTER, NORPSEUDO-B12, TETRACHLOROETHENE REDUCTIVE DEHALOGENASE CATALYTIC SUBUNIT | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UR0
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with trichloroethene | | Descriptor: | 1,1,2-trichloroethene, BENZAMIDINE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
7O7A
 
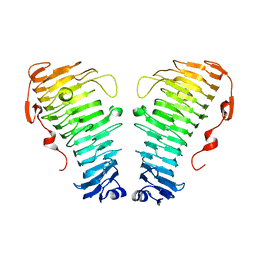 | |
4UXJ
 
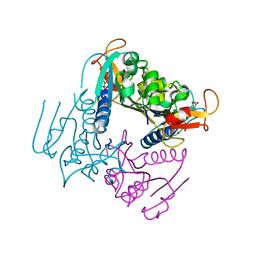 | | Leishmania major Thymidine Kinase in complex with dTTP | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
7O78
 
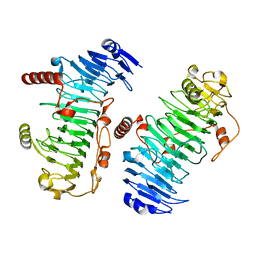 | | Structure of the PL6 family chondroitinase B from Pseudopedobacter saltans, Pedsa3807 | | Descriptor: | Polysaccharide lyase from Pseudopedobacter saltans, Pedsa3807 | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
4UTJ
 
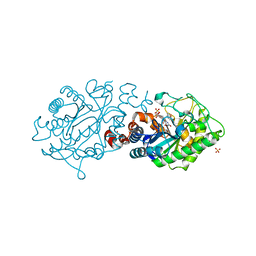 | |
7NUG
 
 | |
4UTQ
 
 | | A structural model of the active ribosome-bound membrane protein insertase YidC | | Descriptor: | ATP SYNTHASE SUBUNIT C, MEMBRANE PROTEIN INSERTASE YIDC | | Authors: | Wickles, S, Singharoy, A, Andreani, J, Seemayer, S, Bischoff, L, Berninghausen, O, Soeding, J, Schulten, K, vanderSluis, E.O, Beckmann, R. | | Deposit date: | 2014-07-22 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A Structural Model of the Active Ribosome-Bound Membrane Protein Insertase Yidc.
Elife, 3, 2014
|
|
7O79
 
 | | Structure of the PL6 family polysaccharide lyase Pedsa3628 from Pseudopedobacter saltans | | Descriptor: | PHOSPHATE ION, Poly(Beta-D-mannuronate) lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
8CWV
 
 | |
7NUH
 
 | |
1BDI
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Glasfeld, A, Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1996-07-25 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Positively Charged Residue Bound in the Minor Groove Does not Alter the Bending of a DNA Duplex
J.Am.Chem.Soc., 118, 1996
|
|
4USN
 
 | | The structure of the immature HIV-1 capsid in intact virus particles at sub-nm resolution | | Descriptor: | P24 | | Authors: | Schur, F.K.M, Hagen, W.J.H, Rumlova, M, Ruml, T, Mueller, B, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structure of the Immature HIV-1 Capsid in Intact Virus Particles at 8.8 A Resolution.
Nature, 517, 2015
|
|
8CWU
 
 | |
4UXT
 
 | | Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, KINESIN HEAVY CHAIN ISOFORM 5A, ... | | Authors: | Atherton, J, Farabella, I, Yu, I.M, Rosenfeld, S.S, Houdusse, A, Topf, M, Moores, C. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Conserved Mechanisms of Microtubule-Stimulated Adp Release, ATP Binding, and Force Generation in Transport Kinesins.
Elife, 3, 2014
|
|
