4GWT
 
 | | Structure of racemic Pin1 WW domain cocrystallized with DL-malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2CF9
 
 | | Complex of recombinant human thrombin with an inhibitor | | Descriptor: | 4-[(1R,3AS,4R,8AS,8BR)-1-ISOPROPYL-2-(4-METHOXYBENZYL)-3-OXODECAHYDROPYRROLO[3,4-A]PYRROLIZIN-4-YL]BENZENECARBOXIMIDAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Schweizer, E, Hoffmann-Roeder, A, Olsen, J.A, Obst-Sander, U, Wagner, B, Kansy, M, Banner, D.W, Diederich, F. | | Deposit date: | 2006-02-17 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Multipolar Interactions in the D Pocket of Thrombin: Large Differences between Tricyclic Imide and Lactam Inhibitors.
Org.Biomol.Chem., 4, 2006
|
|
3PP1
 
 | | Crystal Structure of the Human Mitogen-activated protein kinase kinase 1 (MEK 1) in complex with ligand and MgATP | | Descriptor: | 3-[(2R)-2,3-dihydroxypropyl]-6-fluoro-5-[(2-fluoro-4-iodophenyl)amino]-8-methylpyrido[2,3-d]pyrimidine-4,7(3H,8H)-dione, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2010-11-23 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4GUE
 
 | | Structure of N-terminal kinase domain of RSK2 with flavonoid glycoside quercitrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, MAGNESIUM ION, Ribosomal protein S6 kinase alpha-3, ... | | Authors: | Derewenda, U, Utepbergenov, D, Szukalska, G, Derewenda, Z.S. | | Deposit date: | 2012-08-29 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of quercitrin as an inhibitor of the p90 S6 ribosomal kinase (RSK): structure of its complex with the N-terminal domain of RSK2 at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GXG
 
 | | Crystal structure of human GLTP bound with 12:0 monosulfatide (orthorhombic form; four subunits in asymmetric unit) | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Goni-de-Cerio, F, Popov, A.N, Malinina, L. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3W5P
 
 | | Crystal structure of complexes of vitamin D receptor ligand binding domain with lithocholic acid derivatives | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Masuno, H, Ikura, T, Ito, N. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of complexes of vitamin D receptor ligand-binding domain with lithocholic acid derivatives.
J.Lipid Res., 54, 2013
|
|
4GWV
 
 | | Structure of racemic Pin1 WW domain cocrystallized with tri-ammonium citrate | | Descriptor: | CITRATE ANION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Yun, H.G, Gellman, S.H, Forest, K.T. | | Deposit date: | 2012-09-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Evidence for small-molecule-mediated loop stabilization in the structure of the isolated Pin1 WW domain.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1OI6
 
 | | Structure determination of the TMP-complex of EvaD | | Descriptor: | GLYCEROL, PCZA361.16, THYMIDINE-5'-PHOSPHATE | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-06-09 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Position of a Key Tyrosine in Dtdp-4-Keto-6-Deoxy-D-Glucose-5-Epimerase (Evad) Alters the Substrate Profile for This Rmlc-Like Enzyme
J.Biol.Chem., 279, 2004
|
|
3RWQ
 
 | | Discovery of a Novel, Potent and Selective Inhibitor of 3-Phosphoinositide Dependent Kinase (PDK1) | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Kazmirski, S, Kohls, D. | | Deposit date: | 2011-05-09 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Novel, Potent, and Selective Inhibitors of 3-Phosphoinositide-Dependent Kinase (PDK1).
J.Med.Chem., 54, 2011
|
|
1ZPH
 
 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8315 complexed with CGCGAATTCGCG | | Descriptor: | 1,6-DIMETHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1CF2
 
 | | THREE-DIMENSIONAL STRUCTURE OF D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM THE HYPERTHERMOPHILIC ARCHAEON METHANOTHERMUS FERVIDUS | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), SULFATE ION | | Authors: | Charron, C, Talfournier, F, Isuppov, M.N, Branlant, G, Littlechild, J.A, Vitoux, B, Aubry, A. | | Deposit date: | 1999-03-24 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallization and preliminary X-ray diffraction studies of D-glyceraldehyde-3-phosphate dehydrogenase from the hyperthermophilic archaeon Methanothermus fervidus.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2VVP
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
2VZT
 
 | | Complex of Amycolatopsis orientalis exo-chitosanase CsxA E541A with PNP-beta-D-glucosamine | | Descriptor: | 4-nitrophenyl 2-amino-2-deoxy-beta-D-glucopyranoside, ACETATE ION, CADMIUM ION, ... | | Authors: | Lammerts van Bueren, A, Ghinet, M.G, Gregg, K, Fleury, A, Brzezinski, R, Boraston, A.B. | | Deposit date: | 2008-08-05 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Substrate Recognition in an Exo-Beta-D-Glucosaminidase Involved in Chitosan Hydrolysis.
J.Mol.Biol., 385, 2009
|
|
2OST
 
 | | The structure of a bacterial homing endonuclease : I-Ssp6803I | | Descriptor: | CALCIUM ION, Putative endonuclease, Synthetic DNA 29 MER | | Authors: | Zhao, L, Bonocora, R.P, Shub, D.A, Stoddard, B.L. | | Deposit date: | 2007-02-06 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif.
Embo J., 26, 2007
|
|
5YNA
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM alpha-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1DFG
 
 | | X-RAY STRUCTURE OF ESCHERICHIA COLI ENOYL REDUCTASE WITH BOUND NAD AND BENZO-DIAZABORINE | | Descriptor: | 2-(TOLUENE-4-SULFONYL)-2H-BENZO[D][1,2,3]DIAZABORININ-1-OL, ENOYL ACYL CARRIER PROTEIN REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Baldock, C, Rafferty, J.B, Rice, D.W. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mechanism of drug action revealed by structural studies of enoyl reductase.
Science, 274, 1996
|
|
5YNC
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 1 mM beta-cyclodextrin | | Descriptor: | ACETATE ION, CALCIUM ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4MOL
 
 | | Pyranose 2-oxidase H167A mutant with 2-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-galactopyranose, DODECAETHYLENE GLYCOL, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
2B3E
 
 | | Crystal structure of DB819-D(CGCGAATTCGCG)2 complex. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', 6-(4,5-DIHYDRO-1H-IMIDAZOL-2-YL)-2-{5-[4-(4,5-DIHYDRO-1H-IMIDAZOL-2-YL)PHENYL]THIEN-2-YL}-1H-BENZIMIDAZOLE, MAGNESIUM ION | | Authors: | Campbell, N.H, Evans, D.A, Lee, M.P, Parkinson, G.N, Neidle, S. | | Deposit date: | 2005-09-20 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Targeting the DNA minor groove with fused ring dicationic compounds: Comparison of in silico screening and a high-resolution crystal structure.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4MOJ
 
 | | Pyranose 2-oxidase H450G/V546C double mutant with 2-fluorinated glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOQ
 
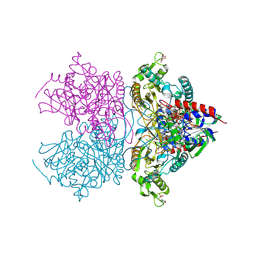 | | Pyranose 2-oxidase V546C mutant with 2-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOP
 
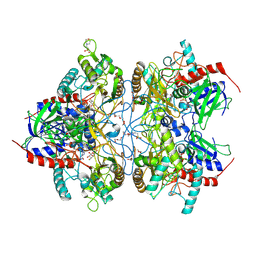 | | Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MIH
 
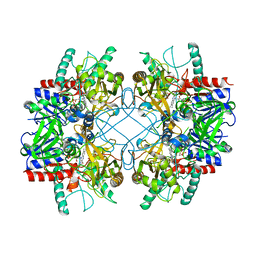 | | Pyranose 2-oxidase from Phanerochaete chrysosporium, recombinant H158A mutant | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Hassan, N, Tan, T.C, Spadiut, O, Pisanelli, I, Fusco, L, Haltrich, D, Peterbauer, C, Divne, C. | | Deposit date: | 2013-08-31 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of Phanerochaete chrysosporium pyranose 2-oxidase suggest that the N-terminus acts as a propeptide that assists in homotetramer assembly.
FEBS Open Bio, 3, 2013
|
|
4ZN6
 
 | |
2ATM
 
 | | Crystal structure of the recombinant allergen Ves v 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hyaluronoglucosaminidase, SULFATE ION | | Authors: | Skov, L.K, Seppala, U, Coen, J.J.F, Crickmore, N, King, T.P, Monsalve, R, Kastrup, J.S, Spangfort, M.D, Gajhede, M. | | Deposit date: | 2005-08-25 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant Ves v 2 at 2.0 Angstrom resolution: structural analysis of an allergenic hyaluronidase from wasp venom.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
