6O7V
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OQW
 
 | | E. coli ATP synthase State 3a | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6O7W
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 2 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OQS
 
 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OQT
 
 | | E. coli ATP synthase State 1c | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7XKH
 
 | | Nucleotide-depleted F1 domain of FoF1-ATPase from Bacillus PS3, state1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKO
 
 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,nucleotide depeleted | | Descriptor: | ATP synthase gamma chain, ATP synthase subunit alpha, ATP synthase subunit beta, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKQ
 
 | | F1 domain of FoF1-ATPase with the down form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKR
 
 | | F1 domain of FoF1-ATPase with the up form of epsilon subunit from Bacillus PS3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
7XKP
 
 | | F1 domain of epsilon C-terminal domain deleted FoF1 from Bacillus PS3,state1,unisite condition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Nakano, A, Kishikawa, J, Nakanishi, A, Mitsuoka, K, Yokoyama, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of unisite catalysis of bacterial F 0 F 1 -ATPase.
Pnas Nexus, 1, 2022
|
|
6YNY
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite dimer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6XBW
 
 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
6PQV
 
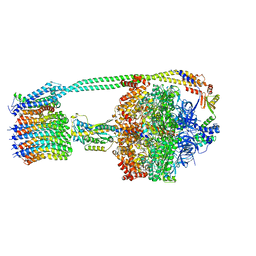 | | E. coli ATP Synthase State 1e | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-07-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
7JG7
 
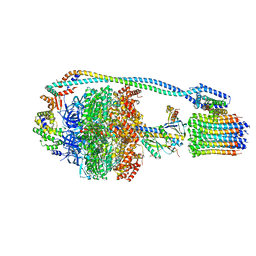 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
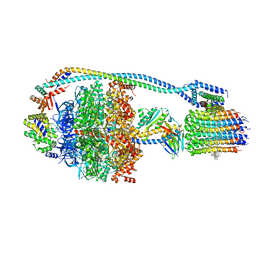 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
6WM3
 
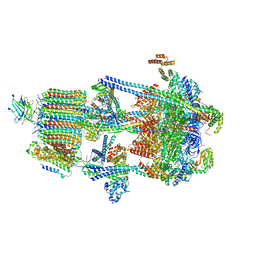 | | Human V-ATPase in state 2 with SidK and ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG6
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
6WM2
 
 | | Human V-ATPase in state 1 with SidK and ADP | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, L, Wu, H, Fu, T.M. | | Deposit date: | 2020-04-20 | | Release date: | 2020-11-11 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
7Y5C
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5A
 
 | | Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Wong, C.F, Saw, W.-G, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5D
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
6YO0
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1/peripheral stalk | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
6YNZ
 
 | | Cryo-EM structure of Tetrahymena thermophila mitochondrial ATP synthase - F1Fo composite tetramer model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kock Flygaard, R, Muhleip, A, Amunts, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Type III ATP synthase is a symmetry-deviated dimer that induces membrane curvature through tetramerization.
Nat Commun, 11, 2020
|
|
