3I4L
 
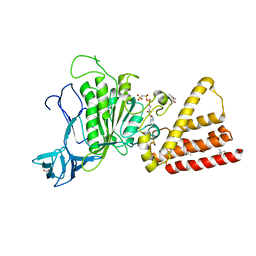 | | Structural characterization for the nucleotide binding ability of subunit A with AMP-PNP of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | Authors: | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3J9T
 
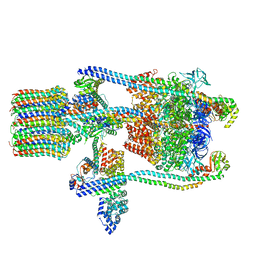 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
5VOY
 
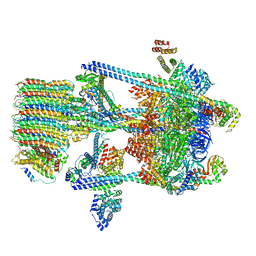 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 2) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
7KHR
 
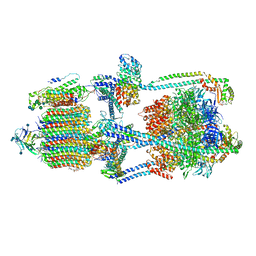 | | Cryo-EM structure of bafilomycin A1-bound intact V-ATPase from bovine brain | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis of V-ATPase inhibition by bafilomycin A1.
Nat Commun, 12, 2021
|
|
3ND9
 
 | |
3P20
 
 | | Crystal structure of vanadate bound subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-10-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
7L1S
 
 | | PS3 F1-ATPase Pi-bound Dwell | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1Q
 
 | | PS3 F1-ATPase Binding/TS Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
7L1R
 
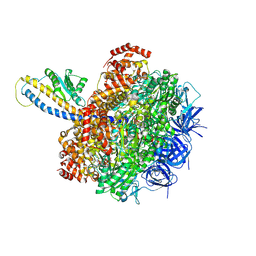 | | PS3 F1-ATPase Hydrolysis Dwell | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Sobti, M, Ueno, H, Noji, H, Stewart, A.G. | | Deposit date: | 2020-12-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Nat Commun, 12, 2021
|
|
4B2Q
 
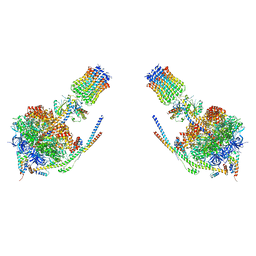 | |
5ZE9
 
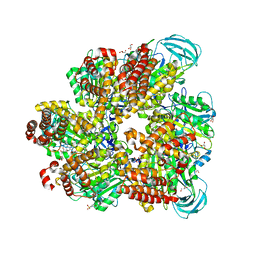 | | Crystal structure of AMP-PNP bound mutant A3B3 complex from Enterococcus hirae V-ATPase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Maruyama, S, Suzuki, K, Sasaki, H, Mizutani, K, Saito, Y, Imai, F.L, Ishizuka-Katsura, Y, Shirouzu, M, Ichiro, Y, Murata, T. | | Deposit date: | 2018-02-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Metastable asymmetrical structure of a shaftless V1motor.
Sci Adv, 5, 2019
|
|
3OE7
 
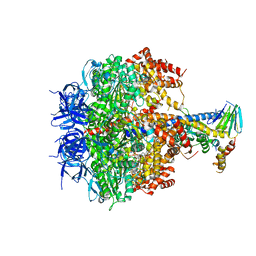 | | Structure of four mutant forms of yeast f1 ATPase: gamma-I270T | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
3W3A
 
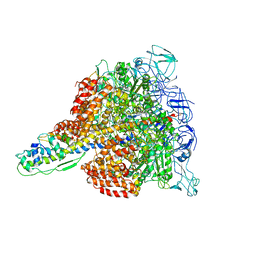 | | Crystal structure of V1-ATPase at 3.9 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | Authors: | Nagamatsu, Y, Takeda, K, Kuranaga, T, Numoto, N, Miki, K. | | Deposit date: | 2012-12-14 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Origin of Asymmetry at the Intersubunit Interfaces of V1-ATPase from Thermusthermophilus
J.Mol.Biol., 425, 2013
|
|
3OEE
 
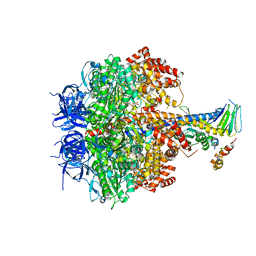 | | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of four mutant forms of yeast F1 ATPase: alpha-F405S
To be Published
|
|
4ASU
 
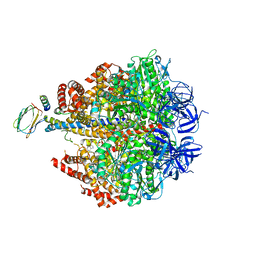 | | F1-ATPase in which all three catalytic sites contain bound nucleotide, with magnesium ion released in the Empty site | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA, MITOCHONDRIAL, ... | | Authors: | Rees, D.M, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2012-05-03 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Evidence of a New Catalytic Intermediate in the Pathway of ATP Hydrolysis by F1-ATPase from Bovine Heart Mitochondria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OFN
 
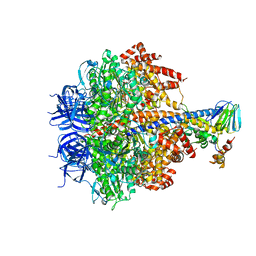 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
8APF
 
 | | rotational state 2a of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APD
 
 | | rotational state 1d of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APH
 
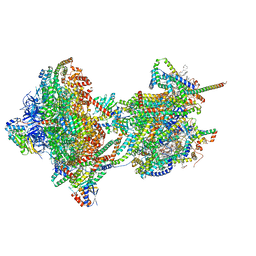 | | rotational state 2c of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APJ
 
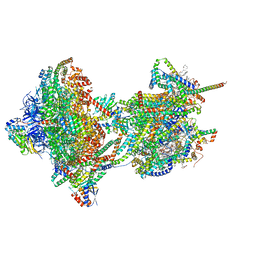 | | rotational state 2d of Trypanosoma brucei mitochondrial ATP synthase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APC
 
 | | rotational state 1c of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APB
 
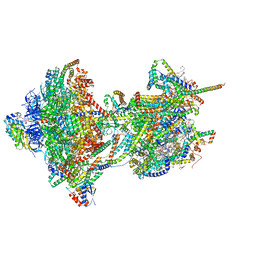 | | rotational state 1b of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APE
 
 | | rotational state 1e of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APA
 
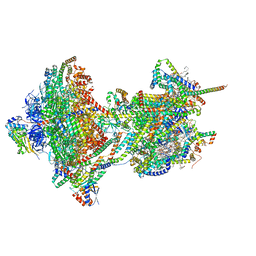 | | rotational state 1a of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8APK
 
 | | rotational state 3 of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
