8RZD
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-(3-hydroxyphenyl)benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 9
To Be Published
|
|
1F85
 
 | | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIIE | | Descriptor: | HCV-1B IRES RNA DOMAIN IIIE | | Authors: | Lukavsky, P.J, Otto, G.A, Lancaster, A.M, Sarnow, P, Puglisi, J.D. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function.
Nat.Struct.Biol., 7, 2000
|
|
8RZE
 
 | | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10 | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanylmethyl]-5-pyridin-3-yl-benzoic acid, ... | | Authors: | Kalnins, G. | | Deposit date: | 2024-02-12 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 nsp16-nsp10 in complex with SAM derivative inhibitor 10
To Be Published
|
|
1BSH
 
 | |
1WMH
 
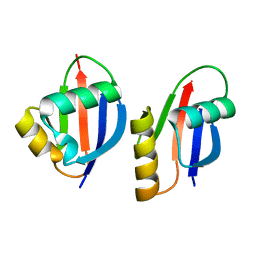 | | Crystal structure of a PB1 domain complex of Protein kinase c iota and Par6 alpha | | Descriptor: | Partitioning defective-6 homolog alpha, Protein kinase C, iota type | | Authors: | Hirano, Y, Yoshinaga, S, Suzuki, N.N, Horiuchi, M, Kohjima, M, Takeya, R, Sumimoto, H, Inagaki, F. | | Deposit date: | 2004-07-09 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a Cell Polarity Regulator, a Complex between Atypical PKC and Par6 PB1 Domains
J.Biol.Chem., 280, 2005
|
|
6BCA
 
 | |
5ME6
 
 | | Crystal Structure of eiF4E from C. melo bound to a CAP analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic transcription initiation factor 4E | | Authors: | Querol-Audi, J, Silva, C, Miras, M, Aranda-Regules, M, Verdaguer, N. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eIF4E in Complex with an eIF4G Peptide Supports a Universal Bipartite Binding Mode for Protein Translation.
Plant Physiol., 174, 2017
|
|
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BWK
 
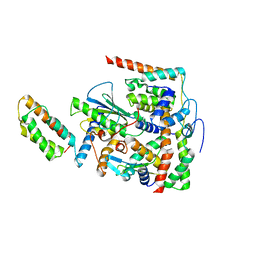 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA-VpdB(461-590) derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Oh, B.H. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex.
Nat Commun, 11, 2020
|
|
6MJH
 
 | |
6SUR
 
 | | The Rab33B-Atg16L1 crystal structure | | Descriptor: | Autophagy-related protein 16-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Metje-Sprink, J, Kuehnel, K. | | Deposit date: | 2019-09-16 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.467 Å) | | Cite: | Crystal structure of the Rab33B/Atg16L1 effector complex.
Sci Rep, 10, 2020
|
|
5J5P
 
 | | AMP-PNP-stabilized ATPase domain of topoisomerase IV from Streptococcus pneumoniae, complex type I | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*C)-3'), DNA topoisomerase 4 subunit B, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Pan, X.-S, Skamrova, G, Umrekar, T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2016-04-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase.
Nat Commun, 9, 2018
|
|
8A8F
 
 | | Crystal structure of Glc7 phosphatase in complex with the regulatory region of Ref2 | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, RNA end formation protein 2, ... | | Authors: | Carminati, M, Manav, C.M, Bellini, D, Passmore, L.A. | | Deposit date: | 2022-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A direct interaction between CPF and RNA Pol II links RNA 3' end processing to transcription.
Mol.Cell, 83, 2023
|
|
1BTM
 
 | | TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH 2-PHOSPHOGLYCOLIC ACID | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Delboni, L.F, Mande, S.C, Hol, W.G.J. | | Deposit date: | 1995-11-11 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of recombinant triosephosphate isomerase from Bacillus stearothermophilus. An analysis of potential thermostability factors in six isomerases with known three-dimensional structures points to the importance of hydrophobic interactions.
Protein Sci., 4, 1995
|
|
5M76
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H10 | | Descriptor: | BROMIDE ION, light chain dimer | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-26 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
6AVU
 
 | | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | | Descriptor: | Fab LM609 heavy chain, Fab LM609 light chain, Integrin alpha-V, ... | | Authors: | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
7KJH
 
 | | Plasmodium falciparum protein Pf12p bound to nanobody B9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Nanobody B9, ... | | Authors: | Dietrich, M.H, Tham, W.H. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nanobody generation and structural characterization of Plasmodium falciparum 6-cysteine protein Pf12p.
Biochem.J., 478, 2021
|
|
8G5C
 
 | |
5IUX
 
 | | GLIC-V135C bimane labelled X-ray structure | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2,3,5,6-tetramethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, ACETATE ION, ... | | Authors: | Fourati, Z, Menny, A, Delarue, M. | | Deposit date: | 2016-03-18 | | Release date: | 2017-03-29 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a pre-active conformation of a pentameric channel receptor.
Elife, 6, 2017
|
|
7KMS
 
 | | Cryo-EM structure of triple ACE2-bound SARS-CoV-2 trimer spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6IYC
 
 | | Recognition of the Amyloid Precursor Protein by Human gamma-secretase | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, R, Yang, G, Guo, X, Zhou, Q, Lei, J, Shi, Y. | | Deposit date: | 2018-12-14 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of the amyloid precursor protein by human gamma-secretase.
Science, 363, 2019
|
|
1FIA
 
 | | CRYSTAL STRUCTURE OF THE FACTOR FOR INVERSION STIMULATION FIS AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FACTOR FOR INVERSION STIMULATION (FIS) | | Authors: | Kostrewa, D, Granzin, J, Choe, H.-W, Labahn, J, Saenger, W. | | Deposit date: | 1991-12-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the factor for inversion stimulation FIS at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
5J5G
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 6-(4-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5MCV
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC1) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
