3V9H
 
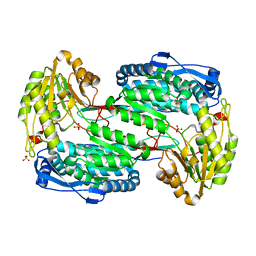 | |
2VE1
 
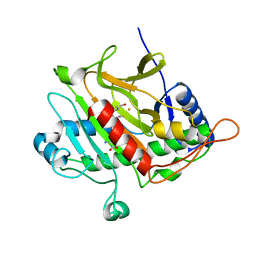 | | Isopenicillin N synthase with substrate analogue AsMCOV (oxygen exposed 1min 20bar) | | Descriptor: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | Authors: | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2007-10-15 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
4FGH
 
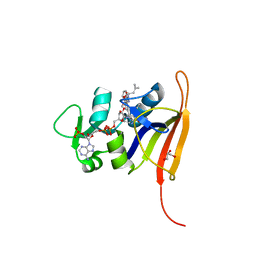 | | S. aureus dihydrofolate reductase co-crystallized with ethyl-DAP isobutenyl-dihydrophthalazine inhibitor | | Descriptor: | (2E)-3-{5-[(2,4-diamino-6-ethylpyrimidin-5-yl)methyl]-2,3-dimethoxyphenyl}-1-[(1S)-1-(2-methylprop-1-en-1-yl)phthalazin-2(1H)-yl]prop-2-en-1-one, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Bourne, C.R, Barrow, W.W. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of Bacterial Dihydrofolate Reductase by 6-Alkyl-2,4-diaminopyrimidines.
Chemmedchem, 7, 2012
|
|
3AM9
 
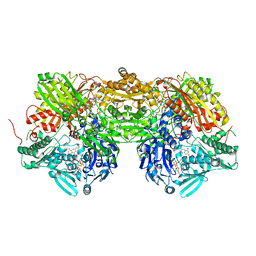 | | Complex of bovine xanthine dehydrogenase and trihydroxy FYX-051 | | Descriptor: | 4-[5-(2,6-dioxo-1,2,3,6-tetrahydropyridin-4-yl)-1H-1,2,4-triazol-3-yl]-6-oxo-1,6-dihydropyridine-2-carbonitrile, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Matsumoto, K, Okamoto, K, Ashizawa, N, Matsumura, T, Kusano, T, Nishino, T. | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | FYX-051: A Novel and Potent Hybrid-Type Inhibitor of Xanthine Oxidoreductase
J.Pharmacol.Exp.Ther., 336, 2011
|
|
4N6H
 
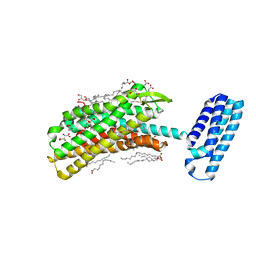 | | 1.8 A Structure of the human delta opioid 7TM receptor (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,14,14b-hexahydro-4,8-methano[1]benzofuro[2,3-a]pyrido[4,3-b]carbazole-1,8a(9H)-diol, L(+)-TARTARIC ACID, ... | | Authors: | Fenalti, G, Giguere, P.M, Katritch, V, Huang, X.-P, Thompson, A.A, Han, G.W, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-12 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular control of delta-opioid receptor signalling.
Nature, 506, 2014
|
|
4H98
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-{3-[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine (UCP1018) | | Descriptor: | 5-{3-[3-(1,3-benzodioxol-5-yl)-5-methoxyphenyl]prop-1-yn-1-yl}-6-ethylpyrimidine-2,4-diamine, Dihydrofolate Reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-09-24 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of the active sites of dihydrofolate reductase from two species of Candida uncovers ligand-induced conformational changes shared among species.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2PDH
 
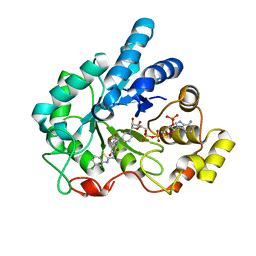 | | Human aldose reductase mutant L300P complexed with uracil-type inhibitor at 1.45 A. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {3-[(5-CHLORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
4NMF
 
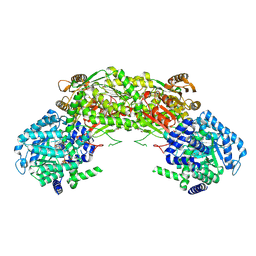 | | Crystal structure of proline utilization A (PutA) from Geobacter sulfurreducens PCA inactivated by N-propargylglycine and complexed with menadione bisulfite | | Descriptor: | (2R)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, (2S)-2-methyl-1,4-dioxo-1,2,3,4-tetrahydronaphthalene-2-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Singh, H, Tanner, J.J. | | Deposit date: | 2013-11-14 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone-binding site.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NEV
 
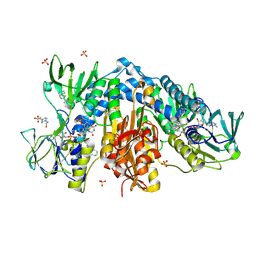 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor EP127 (5-{5-[1-(PYRROLIDIN-1-YL)CYCLOHEXYL]-1,3-THIAZOL-2-YL}-1H-INDOLE) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-2-yl}-1H-indole, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Persch, E, Bryson, S, Pai, E.F, Krauth-Siegel, R.L, Diederich, F. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding to large enzyme pockets: small-molecule inhibitors of trypanothione reductase.
Chemmedchem, 9, 2014
|
|
3RLH
 
 | | Crystal structure of a class II phospholipase D from Loxosceles intermedia venom | | Descriptor: | (2r,5r)-5-amino-2-hydroxy-5-(hydroxymethyl)-1,3,2lambda~5~-dioxaphosphinan-2-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K, Doherty, D.Z, Gismene, C, Bachega, J.F.R, Chahine, J, Gonzalez, J.E.H. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
1IMX
 
 | | 1.8 Angstrom crystal structure of IGF-1 | | Descriptor: | BROMIDE ION, Insulin-like Growth Factor 1A, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE | | Authors: | Vajdos, F.F, Ultsch, M, Schaffer, M.L, Deshayes, K.D, Liu, J, Skelton, N.J, de Vos, A.M. | | Deposit date: | 2001-05-11 | | Release date: | 2001-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions.
Biochemistry, 40, 2001
|
|
3NT4
 
 | | Crystal structure of myo-inositol dehydrogenase from Bacillus subtilis with bound cofactor NADH and inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5001 Å) | | Cite: | Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification.
Biochem.J., 432, 2010
|
|
2PPC
 
 | |
9KRY
 
 | | Crystal structure Of MerTK kinase domain in complex With compound 1 | | Descriptor: | Tyrosine-protein kinase Mer, ~{N}-[4-[5-(3-aminophenyl)-6-(1-methylpyrazol-4-yl)furo[2,3-d]pyrimidin-4-yl]oxyphenyl]-2-(4-fluorophenyl)-1-methyl-3-oxidanylidene-pyrazole-4-carboxamide | | Authors: | Peng, Y.H, Lee, L.C, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2024-11-29 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design of Potent and Selective MerTK Inhibitors by Modulating the Conformation of alpha C Helix.
J.Med.Chem., 68, 2025
|
|
2OHK
 
 | |
4L1O
 
 | | Crystal structure of human ALDH3A1 with inhibitor 1-{[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]methyl}-1H-indole-2,3-dione | | Descriptor: | (3S)-1-{[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]methyl}-3-hydroxy-1,3-dihydro-2H-indol-2-one, ACETATE ION, Aldehyde dehydrogenase, ... | | Authors: | Hurley, T.D, Parajuli, B. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones.
J.Med.Chem., 57, 2014
|
|
1B14
 
 | | Alcohol Dehydrogenase from Drosophila Lebanonensis Binary Complex with NAD+ | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Benach, J, Atrian, S, Gonzalez-Duarte, R, Ladenstein, R. | | Deposit date: | 1998-11-25 | | Release date: | 1999-11-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The catalytic reaction and inhibition mechanism of Drosophila alcohol dehydrogenase: observation of an enzyme-bound NAD-ketone adduct at 1.4 A resolution by X-ray crystallography.
J.Mol.Biol., 289, 1999
|
|
3SBC
 
 | | Crystal structure of Saccharomyces cerevisiae TSA1C47S mutant protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, Peroxiredoxin TSA1 | | Authors: | Tairum Jr, C.A, Horta, B.B, Netto, L.E.S, Oliveira, M.A. | | Deposit date: | 2011-06-03 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disulfide biochemistry in 2-cys peroxiredoxin: requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin.
J.Mol.Biol., 424, 2012
|
|
4FPB
 
 | |
3WIE
 
 | | Structure of a glucose dehydrogenase T277F mutant in complex with D-glucose and NAADP | | Descriptor: | Glucose 1-dehydrogenase, ZINC ION, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(3-carboxypyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate, ... | | Authors: | Sakuraba, H, Kanoh, Y, Yoneda, K, Ohshima, T. | | Deposit date: | 2013-09-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into glucose dehydrogenase from the thermoacidophilic archaeon Thermoplasma volcanium.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1PU8
 
 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
4I4G
 
 | | Crystal structure of CYP3A4 ligated to oxazole-substituted desoxyritonavir | | Descriptor: | Cytochrome P450 3A4, N~2~-(methyl{[2-(propan-2-yl)-1,3-thiazol-4-yl]methyl}carbamoyl)-N-[(2R,5R)-5-{[(1,3-oxazol-5-ylmethoxy)carbonyl]amino}-1,6-diphenylhexan-2-yl]-L-valinamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F, Poulos, T.L. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pyridine-Substituted Desoxyritonavir Is a More Potent Inhibitor of Cytochrome P450 3A4 than Ritonavir.
J.Med.Chem., 56, 2013
|
|
2I7S
 
 | | Crystal structure of Re(phen)(CO)3 (Thr124His)(His83Gln) Azurin Cu(II) from Pseudomonas aeruginosa | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COBALT TETRAAMMINE ION, ... | | Authors: | Gradinaru, C, Crane, B.R. | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Relaxation dynamics of Pseudomonas aeruginosa Re(I)(CO)3(alpha-diimine)(HisX)+ (X = 83, 107, 109, 124, 126)Cu(II) azurins.
J.Am.Chem.Soc., 131, 2009
|
|
3ZOS
 
 | | Structure of the DDR1 kinase domain in complex with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | Authors: | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Bradley, A, Coutandin, D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-02-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Mechanisms Determining Inhibition of the Collagen Receptor Ddr1 by Selective and Multi-Targeted Type II Kinase Inhibitors
J.Mol.Biol., 426, 2014
|
|
