6PLQ
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, super-open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM3
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, closed state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM1
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, desensitized state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM5
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, desensitized state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM4
 
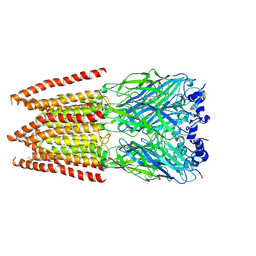 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, super-open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLP
 
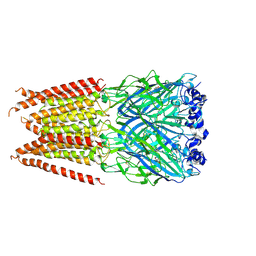 | | CryoEM structure of zebra fish alpha-1 glycine receptor YGF mutant bound with GABA in SMA, desensitized state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM2
 
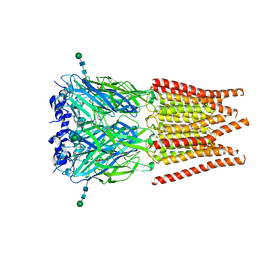 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Taurine in SMA, open state | | Descriptor: | 2-AMINOETHANESULFONIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PM6
 
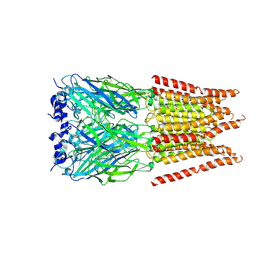 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with Glycine in SMA, open state | | Descriptor: | GLYCINE, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
6PLW
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, super-open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
1GE6
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6PXD
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor, Apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Gouaux, E. | | Deposit date: | 2019-07-25 | | Release date: | 2021-02-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
1G12
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-10 | | Release date: | 2001-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GE7
 
 | | ZINC PEPTIDASE FROM GRIFOLA FRONDOSA | | Descriptor: | PEPTIDYL-LYS METALLOENDOPEPTIDASE, ZINC ION, alpha-D-mannopyranose | | Authors: | Hori, T, Kumasaka, T, Yamamoto, M, Nonaka, T, Tanaka, N, Hashimoto, Y, Ueki, T, Takio, K. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a new 'aspzincin' metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4EII
 
 | |
4XOB
 
 | | Crystal structure of a FimH*DsF complex from E.coli K12 with bound heptyl alpha-D-mannopyrannoside | | Descriptor: | FimF, Protein FimH, SULFATE ION, ... | | Authors: | Jakob, R.P, Eras, J, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
1P5G
 
 | |
5N1I
 
 | |
1PCM
 
 | |
1P5D
 
 | |
3ZBY
 
 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
3V5V
 
 | | UNLIGANDED E.CLOACAE C115D MURA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
4MJ1
 
 | | unliganded BK Polyomavirus VP1 pentamer | | Descriptor: | CHLORIDE ION, GLYCEROL, VP1 capsid protein | | Authors: | Neu, U, Stroh, L.J, Stehle, T. | | Deposit date: | 2013-09-03 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus.
Plos Pathog., 9, 2013
|
|
4OLP
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Pectobacterium wasabiae | | Descriptor: | GrpU microcompartment shell protein | | Authors: | Wheatley, N.M, Thompson, M.C, Gidaniyan, S.D, Sawaya, M.R, Jorda, J, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
4OLO
 
 | | Ligand-free structure of the GrpU microcompartment shell protein from Clostridiales bacterium 1_7_47FAA | | Descriptor: | BMC domain protein | | Authors: | Thompson, M.C, Ahmed, H, McCarty, K.N, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a unique fe-s cluster binding site in a glycyl-radical type microcompartment shell protein.
J.Mol.Biol., 426, 2014
|
|
3SPB
 
 | | Unliganded E. Cloacae MurA | | Descriptor: | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-07-01 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
