3M1V
 
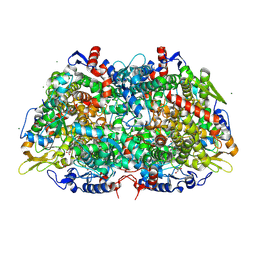 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | Deposit date: | 2010-03-05 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
3M67
 
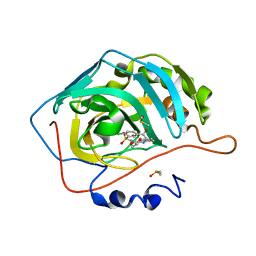 | | Crystal structure of human carbonic anhydrase isozyme II with 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide | | Descriptor: | 2-chloro-5-[(6,7-dihydro-1H-[1,4]dioxino[2,3-f]benzimidazol-2-ylsulfanyl)acetyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2010-03-15 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Indapamide-like benzenesulfonamides as inhibitors of carbonic anhydrases I, II, VII, and XIII
Bioorg.Med.Chem., 18, 2010
|
|
3P3R
 
 | |
3PA5
 
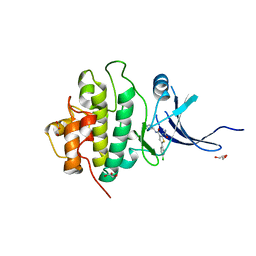 | | X-ray crystal structure of compound 1 bound to human CHK1 kinase domain | | Descriptor: | 2-(carbamoylamino)-5-(4-chlorophenyl)-N-[(3S)-piperidin-3-yl]thiophene-3-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3PB3
 
 | | Structure of an Antibiotic Related Methyltransferase | | Descriptor: | 16S rRNA methylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sivaraman, J, Husain, N. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit.
Nucleic Acids Res., 2010
|
|
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P6F
 
 | |
3OMK
 
 | |
3P6P
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio | | Descriptor: | (3S,5S,7S)-N-[6-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexyl]tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C6-Bio
To be Published
|
|
3OO7
 
 | |
3P7W
 
 | |
3P94
 
 | |
3OOL
 
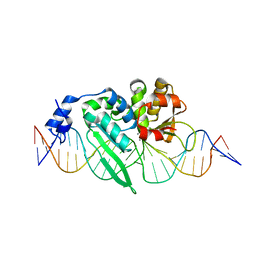 | | I-SceI complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
3P9Q
 
 | | Structure of I274C variant of E. coli KatE | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE 17R, 18S, ... | | Authors: | Loewen, P.C, Jha, V, Louis, S, Chelikani, P, Carpena, X, Fita, I. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Modulation of heme orientation and binding by a single residue in catalase HPII of Escherichia coli.
Biochemistry, 50, 2011
|
|
3OKO
 
 | | Crystal structure of S25-39 in complex with Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
3OPW
 
 | | Crystal Structure of the Rph1 catalytic core | | Descriptor: | DNA damage-responsive transcriptional repressor RPH1 | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
3OQF
 
 | | Crystal Structure Analysis of Renin-indole-piperazine inhibitor complexes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-benzyl-1-phenyl-3-(piperazin-1-ylcarbonyl)-1H-indole, Renin | | Authors: | Bocskei, Z. | | Deposit date: | 2010-09-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery and optimization of a new class of potent and non-chiral indole-3-carboxamide-based renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OMS
 
 | | Putative 3-demethylubiquinone-9 3-methyltransferase, PhnB protein, from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, PhnB protein | | Authors: | Osipiuk, J, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of putative 3-demethylubiquinone-9
3-methyltransferase, PhnB protein, from Bacillus cereus.
To be Published
|
|
3OOX
 
 | |
3OSO
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OQB
 
 | | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110 | | Descriptor: | Oxidoreductase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-02 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative oxidoreductase from Bradyrhizobium japonicum USDA 110
To be Published
|
|
3OS7
 
 | |
3OTD
 
 | | Crystal structure of human tRNAHis guanylyltransferase (Thg1)- NaI derivative | | Descriptor: | IODIDE ION, tRNA(His) guanylyltransferase | | Authors: | Hyde, S.J, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2010-09-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | tRNAHis guanylyltransferase (THG1), a unique 3'-5' nucleotidyl transferase, shares unexpected structural homology with canonical 5'-3' DNA polymerases.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OTN
 
 | |
3OV2
 
 | | Curcumin synthase 1 from Curcuma longa | | Descriptor: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
