5JB3
 
 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-REMOTE conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
5G41
 
 | | Crystal structure of adenylate kinase ancestor 4 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5IW0
 
 | |
6HAM
 
 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
5G40
 
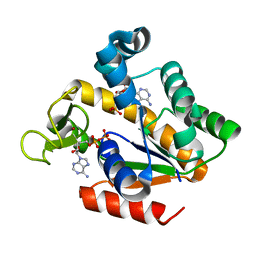 | | Crystal structure of adenylate kinase ancestor 4 with Zn and AMP-ADP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
6KE6
 
 | |
7MPI
 
 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N8B
 
 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | Descriptor: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-06-14 | | Release date: | 2022-05-11 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2ORI
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Shamoo, Y, Wilson, C.J, Wu, G, Myers, J. | | Deposit date: | 2007-02-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (A193V/Q199R/)
To be Published
|
|
8AUV
 
 | | Cryo-EM structure of the plant 40S subunit | | Descriptor: | 18S rRNA, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
2OO7
 
 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Counago, R, Wilson, C.J, Wu, G, Myers, J.C, Wittung-Stafshede, P, Shamoo, Y. | | Deposit date: | 2007-01-25 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (T179I/Q199R)
To be Published
|
|
2OSB
 
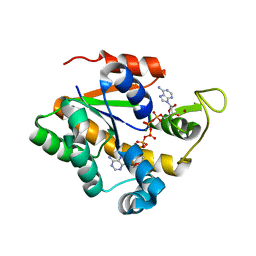 | | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/) | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Myers, J, Counago, R, Wilson, C.J, Wu, G, Shamoo, Y. | | Deposit date: | 2007-02-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a thermostable mutant of Bacillus subtilis Adenylate Kinase (Q16L/Q199R/)
To be Published
|
|
3DL0
 
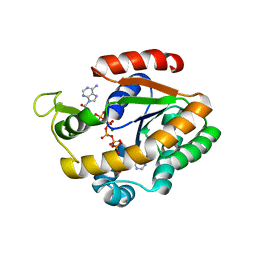 | | Crystal structure of adenylate kinase variant AKlse3 | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, McCoy, J.G. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Effectiveness and limitations of local structural entropy optimization in the thermal stabilization of mesophilic and thermophilic adenylate kinases.
Proteins, 82, 2014
|
|
2AK3
 
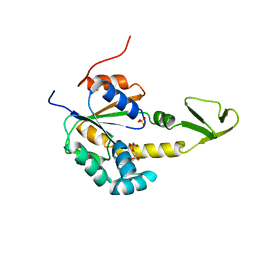 | |
2AK2
 
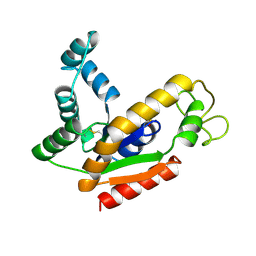 | | ADENYLATE KINASE ISOENZYME-2 | | Descriptor: | ADENYLATE KINASE ISOENZYME-2, SULFATE ION | | Authors: | Schlauderer, G.J, Schulz, G.E. | | Deposit date: | 1995-12-29 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of bovine mitochondrial adenylate kinase: comparison with isoenzymes in other compartments.
Protein Sci., 5, 1996
|
|
3DKV
 
 | | Crystal structure of adenylate kinase variant AKlse1 | | Descriptor: | 1,2-ETHANEDIOL, Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ... | | Authors: | Bannen, R.M, Bianchetti, C.M, Bingman, C.A, Bitto, E.B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of adenylate kinase variant AKlse1.
To be Published
|
|
8C01
 
 | | Enp1TAP_A population of yeast small ribosomal subunit precursors | | Descriptor: | 18S rRNA precursor, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2022-12-15 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Impact of the yeast S0/uS2-cluster ribosomal protein rpS21/eS21 on rRNA folding and the architecture of small ribosomal subunit precursors.
Plos One, 18, 2023
|
|
8C00
 
 | |
8BTD
 
 | | Giardia Ribosome in PRE-T Hybrid State (D1) | | Descriptor: | 5.8S rRNA, 5S rRNA, Large Subunit rRNA, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BSJ
 
 | | Giardia Ribosome in PRE-T Classical State (C) | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Majumdar, S, Emmerich, A.G, Sanyal, S. | | Deposit date: | 2022-11-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.49 Å) | | Cite: | Insights into translocation mechanism and ribosome evolution from cryo-EM structures of translocation intermediates of Giardia intestinalis.
Nucleic Acids Res., 51, 2023
|
|
8BQX
 
 | | Yeast 80S ribosome in complex with Map1 (conformation 2) | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
3UKD
 
 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, CMP, AND ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
5A2N
 
 | |
