8OJY
 
 | |
2RV1
 
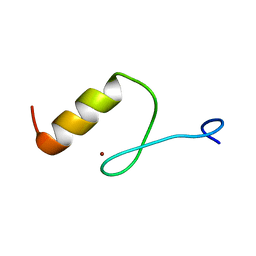 | | Solution structures of the DNA-binding domain (ZF13) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RF7
 
 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
5HW5
 
 | |
5HYB
 
 | |
5HYN
 
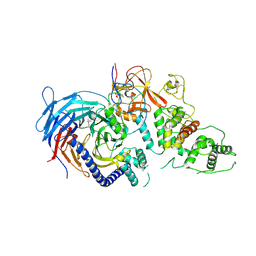 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with oncogenic histone H3K27M peptide | | Descriptor: | H3K27M, Histone-lysine N-methyltransferase EZH2, JARID2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Wilson, J.R, Gamblin, S.J. | | Deposit date: | 2016-02-01 | | Release date: | 2016-05-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Nat Commun, 7, 2016
|
|
5HP1
 
 | |
8OZ5
 
 | |
8P02
 
 | |
2RVA
 
 | |
2RQ8
 
 | | Solution NMR structure of titin I27 domain mutant | | Descriptor: | Titin | | Authors: | Yagawa, K, Oguro, T, Momose, T, Kawano, S, Sato, T, Endo, T. | | Deposit date: | 2009-03-05 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for unfolding pathway-dependent stability of proteins: Vectorial unfolding vs. global unfolding
Protein Sci., 2010
|
|
5HOG
 
 | |
6R0W
 
 | |
5HPL
 
 | |
6R0Z
 
 | |
6R0Y
 
 | |
2RV9
 
 | |
6R15
 
 | | Crystal structure of the SUN1-KASH1 6:6 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nesprin-1, POTASSIUM ION, ... | | Authors: | Gurusaran, M, Davies, O.R. | | Deposit date: | 2019-03-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A molecular mechanism for LINC complex branching by structurally diverse SUN-KASH 6:6 assemblies.
Elife, 10, 2021
|
|
2SCU
 
 | | A detailed description of the structure of Succinyl-COA synthetase from Escherichia coli | | Descriptor: | COENZYME A, PROTEIN (SUCCINYL-COA LIGASE), SULFATE ION | | Authors: | Fraser, M.E, Wolodko, W.T, James, M.N.G, Bridger, W.A. | | Deposit date: | 1998-09-24 | | Release date: | 1999-08-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A detailed structural description of Escherichia coli succinyl-CoA synthetase.
J.Mol.Biol., 285, 1999
|
|
6R10
 
 | |
3QSS
 
 | | Crystal structure for the MSOX.chloride.MTA ternary complex | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
6R2F
 
 | |
6RAA
 
 | | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK1 | | Authors: | Schroeder, M, Arrowsmith, C, Knapp, S, Bountra, C, Edwards, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CLK1 Kinase domain with bound imidazopyridin inhibitor TP003
To Be Published
|
|
2RKY
 
 | |
6RAS
 
 | | Pmar-Lig_Pre. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA, ... | | Authors: | Leiros, H.K.S, Williamson, A. | | Deposit date: | 2019-04-07 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
