7UZW
 
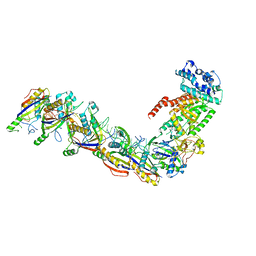 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UZX
 
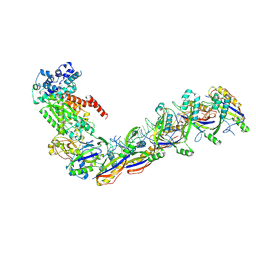 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | Descriptor: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
5ICR
 
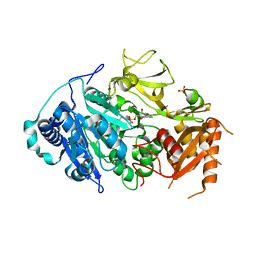 | | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine. | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Acyl-CoA synthase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Hung, D, Fisher, S.L, Edelstein, J, Kiryukhina, O, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-23 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Resolution Crystal Structure of Fatty-Acid-CoA Ligase (FadD32) from Mycobacterium smegmatis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine.
To Be Published
|
|
7ZU9
 
 | |
2YWC
 
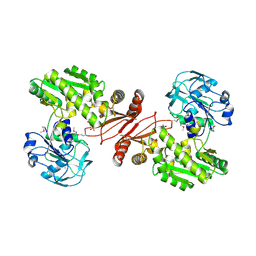 | | Crystal structure of GMP synthetase from Thermus thermophilus in complex with XMP | | Descriptor: | GMP synthase [glutamine-hydrolyzing], XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
2YWB
 
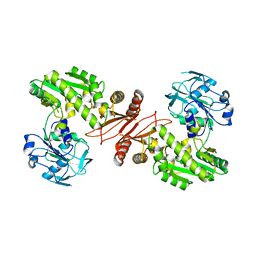 | | Crystal structure of GMP synthetase from Thermus thermophilus | | Descriptor: | GMP synthase [glutamine-hydrolyzing] | | Authors: | Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kuramitsu, S, Yokoyama, S, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of GMP synthetase from Thermus thermophilus
To be Published
|
|
5FLG
 
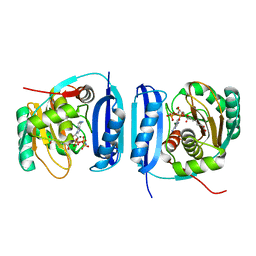 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis in complex with AMPPNP | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5FM0
 
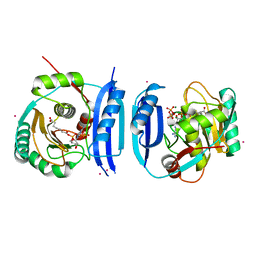 | | Crystal structure of the 6-carboxyhexanoate-CoA ligase (BioW)from Bacillus subtilis (PtCl4 derivative) | | Descriptor: | 6-CARBOXYHEXANOATE--COA LIGASE, MAGNESIUM ION, PIMELOYL-AMP, ... | | Authors: | Moynie, L, Wang, M, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Using the pimeloyl-CoA synthetase adenylation fold to synthesize fatty acid thioesters.
Nat. Chem. Biol., 13, 2017
|
|
5GME
 
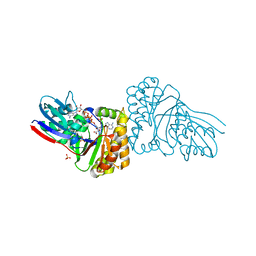 | | Crystal structure of Sulfolobus solfataricus Diphosphomevalonate decarboxylase in complex with ADP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Unno, H, Hemmi, H, Hattori, A. | | Deposit date: | 2016-07-13 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Single Amino Acid Mutation Converts (R)-5-Diphosphomevalonate Decarboxylase into a Kinase
J. Biol. Chem., 292, 2017
|
|
6HXG
 
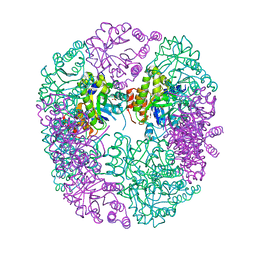 | | PDX1.2/PDX1.3 complex (intermediate) | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1DBS
 
 | |
6SC2
 
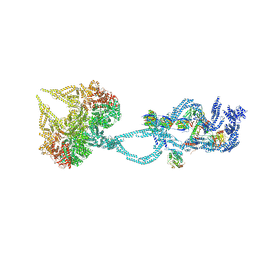 | | Structure of the dynein-2 complex; IFT-train bound model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
6HX3
 
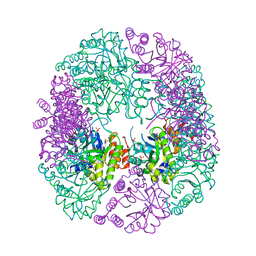 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
3DAY
 
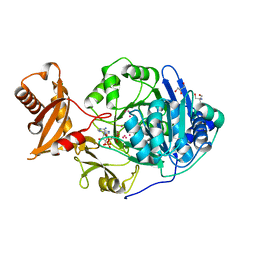 | | Crystal structure of human acyl-CoA synthetase medium-chain family member 2A (L64P mutation) in complex with AMP-CPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-coenzyme A synthetase ACSM2A, mitochondrial precursor, ... | | Authors: | Pilka, E.S, Kochan, G.T, Yue, W.W, Bhatia, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Wikstrom, M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A.
J.Mol.Biol., 388, 2009
|
|
3AT1
 
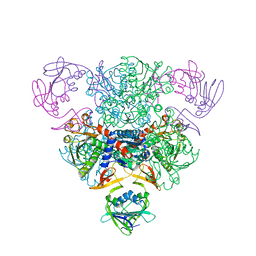 | |
1E19
 
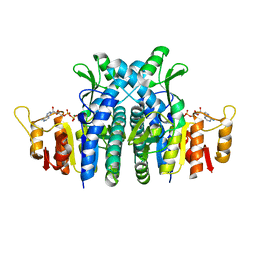 | | Structure of the carbamate kinase-like carbamoyl phosphate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE, MAGNESIUM ION | | Authors: | Ramon-Maiques, S, Marina, A, Uriarte, M, Fita, I, Rubio, V. | | Deposit date: | 2000-04-28 | | Release date: | 2000-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A Resolution Crystal Structure of the Carbamate Kinase-Like Carbamoyl Phosphate Synthetase from the Hyperthermophilic Archaeon Pyrococcus Furiosus, Bound to Adp, Confirms that This Thermoestable Enzyme is a Carbamate Kinase, and Provides Insights Into Substrate Binding and Stability in Carbamate Kinases
J.Mol.Biol., 299, 2000
|
|
3ADC
 
 | | Crystal structure of O-phosphoseryl-tRNA kinase complexed with selenocysteine tRNA and AMPPNP (crystal type 2) | | Descriptor: | L-seryl-tRNA(Sec) kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Itoh, Y, Chiba, S, Sekine, S, Yokoyama, S. | | Deposit date: | 2010-01-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for the Major Role of O-Phosphoseryl-tRNA Kinase in the UGA-Specific Encoding of Selenocysteine
Mol.Cell, 39, 2010
|
|
1WL8
 
 | |
7M4S
 
 | |
6E0M
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase | | Descriptor: | DIPHOSPHATE, cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0N
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6ILD
 
 | | Crystal Structure of Human LysRS: P38/AIMP2 Complex II | | Descriptor: | 5'-O-[(S)-hydroxy(methyl)phosphoryl]adenosine, Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, ... | | Authors: | Hei, Z, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2018-10-17 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex.
J. Biol. Chem., 294, 2019
|
|
3ADD
 
 | | Crystal structure of O-phosphoseryl-tRNA kinase complexed with selenocysteine tRNA and AMPPNP (crystal type 3) | | Descriptor: | L-seryl-tRNA(Sec) kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Itoh, Y, Chiba, S, Sekine, S, Yokoyama, S. | | Deposit date: | 2010-01-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Major Role of O-Phosphoseryl-tRNA Kinase in the UGA-Specific Encoding of Selenocysteine
Mol.Cell, 39, 2010
|
|
3AW8
 
 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Okada, K, Tsunoda, S, Taka, H, Baba, S, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-15 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
