3BTV
 
 | |
4KHB
 
 | | Structure of the Spt16D Pob3N heterodimer | | Descriptor: | SULFATE ION, Uncharacterized protein POB3N, Uncharacterized protein SPT16D | | Authors: | Stuwe, T, Zhang, E, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
5B03
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3BXF
 
 | | Crystal structure of effector binding domain of central glycolytic gene regulator (CggR) from Bacillus subtilis in complex with effector fructose-1,6-bisphosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 1,6-di-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, ... | | Authors: | Rezacova, P, Otwinowski, Z. | | Deposit date: | 2008-01-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the effector-binding domain of repressor Central glycolytic gene Regulator from Bacillus subtilis reveal ligand-induced structural changes upon binding of several glycolytic intermediates.
Mol.Microbiol., 69, 2008
|
|
1B44
 
 | | CRYSTAL STRUCTURE OF THE B SUBUNIT OF HEAT-LABILE ENTEROTOXIN FROM E. COLI CARRYING A PEPTIDE WITH ANTI-HSV ACTIVITY | | Descriptor: | PROTEIN (B-POL SUBUNIT OF HEAT-LABILE ENTEROTOXIN) | | Authors: | Matkovic-Calogovic, D, Loregian, A, D'Acunto, M.R, Battistutta, R, Tossi, A, Palu, G, Zanotti, G. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the B subunit of Escherichia coli heat-labile enterotoxin carrying peptides with anti-herpes simplex virus type 1 activity.
J.Biol.Chem., 274, 1999
|
|
3BXG
 
 | |
4I2P
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) based analogue | | Descriptor: | (2E)-3-[4-({6-[(4-methoxyphenyl)amino]-7H-purin-2-yl}amino)-3,5-dimethylphenyl]prop-2-enenitrile, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2964 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
3BTU
 
 | |
5DGE
 
 | | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Coping with proline stalling: structural basis of hypusine-induced protein synthesis by the eukaryotic ribosome
To Be Published
|
|
3BXE
 
 | |
3BTS
 
 | |
6KTB
 
 | | Crystal structure of B. halodurans MntR in apo form | | Descriptor: | HTH-type transcriptional regulator MntR, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lee, J.Y, Lee, M.Y. | | Deposit date: | 2019-08-26 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the manganese transport regulator MntR from Bacillus halodurans in apo and manganese bound forms.
Plos One, 14, 2019
|
|
1DI2
 
 | |
4JOI
 
 | | Crystal structure of the human telomeric Stn1-Ten1 complex | | Descriptor: | CST complex subunit STN1, CST complex subunit TEN1 | | Authors: | Bryan, C, Rice, C, Harkisheimer, M, Schultz, D, Skordalakes, E. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the human telomeric stn1-ten1 capping complex.
Plos One, 8, 2013
|
|
3U33
 
 | |
2QVA
 
 | |
3QMC
 
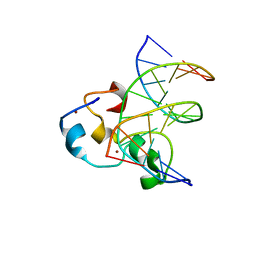 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*CP*TP*GP*GP*C)-3', 5'-D(*GP*CP*CP*AP*GP*CP*GP*GP*TP*GP*GP*C)-3', CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3QMB
 
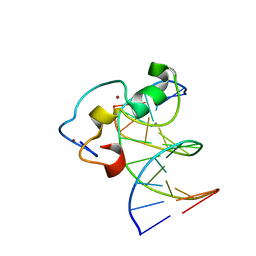 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3', CALCIUM ION, CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3F9V
 
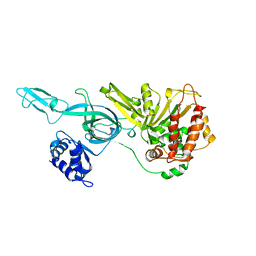 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8IMH
 
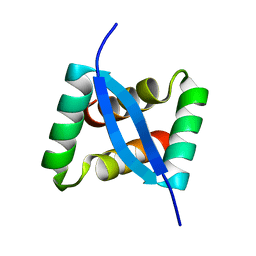 | |
2INR
 
 | |
6BD0
 
 | | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex) | | Descriptor: | CALCIUM ION, DNA (25-MER), Ribosomal protein 3/homing endonuclease-like protein fusion | | Authors: | Brown, C, Zhang, K, Laforet, M, McMurrough, T.A, Gloor, G.B, Edgell, D.R, Junop, M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | I-OnuI K227Y, D236A bound to cognate substrate (pre-cleavage complex)
To Be Published
|
|
6B19
 
 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | Authors: | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | Deposit date: | 2017-09-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
4MMG
 
 | | crystal structure of YafQ mutant H87Q from E.coli | | Descriptor: | SULFATE ION, mRNA interferase YafQ | | Authors: | Liang, Y.J, Gao, Z.Q, Liu, Q.S, Dong, Y.H. | | Deposit date: | 2013-09-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of Escherichia coli Toxin-Antitoxin Complex DinJ-YafQ
J.Biol.Chem., 289, 2014
|
|
1K99
 
 | |
