5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6XIM
 
 | |
6SPW
 
 | | Structure of protein kinase CK2 catalytic subunit with the CK2beta-competitive bisubstrate inhibitor ARC3140 | | Descriptor: | 8-[4,5,6,7-tetrakis(iodanyl)benzimidazol-1-yl]octanoic acid, ARC3140, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Schnitzler, A. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Unexpected CK2 beta-antagonistic functionality of bisubstrate inhibitors targeting protein kinase CK2.
Bioorg.Chem., 96, 2020
|
|
5NN6
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
6SWC
 
 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
7F64
 
 | | eIF2B-SFSV NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Kashiwagi, K, Ito, T. | | Deposit date: | 2021-06-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Nat Commun, 12, 2021
|
|
6SKO
 
 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
1BNQ
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (R)-4-ETHYLAMINO-3,4-DIHYDRO-2-(2-METHOYLETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
5NN4
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
1BNV
 
 | | CARBONIC ANHYDRASE II INHIBITOR | | Descriptor: | (S)-3,4-DIHYDRO-2-(3-METHOXYPHENYL)-4-METHYLAMINO-2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE-1,1-DIOXIDE, CARBONIC ANHYDRASE, MERCURY (II) ION, ... | | Authors: | Boriack-Sjodin, P.A, Zeitlin, S, Christianson, D.W. | | Deposit date: | 1998-07-30 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of inhibitor binding to human carbonic anhydrase II.
Protein Sci., 7, 1998
|
|
3O2K
 
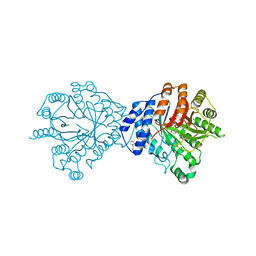 | | Crystal Structure of Brevianamide F Prenyltransferase Complexed with Brevianamide F and Dimethylallyl S-thiolodiphosphate | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Brevianamide F prenyltransferase, ... | | Authors: | Jost, M, Zocher, G.E, Stehle, T. | | Deposit date: | 2010-07-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-function analysis of an enzymatic prenyl transfer reaction identifies a reaction chamber with modifiable specificity.
J.Am.Chem.Soc., 132, 2010
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
4UFR
 
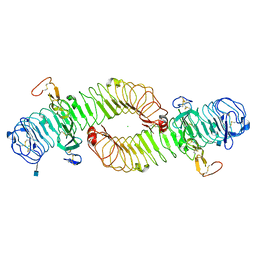 | |
6EMK
 
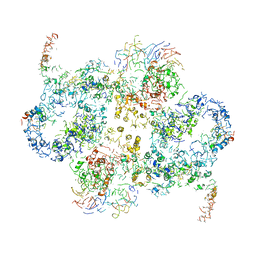 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
6ALJ
 
 | | ALDH1A2 liganded with NAD and compound WIN18,446 | | Descriptor: | Aldehyde dehydrogenase 1A2, N,N'-(octane-1,8-diyl)bis(2,2-dichloroacetamide), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chen, Y, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-08-08 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of ALDH1A2 Inhibition by Irreversible and Reversible Small Molecule Inhibitors.
ACS Chem. Biol., 13, 2018
|
|
7WJU
 
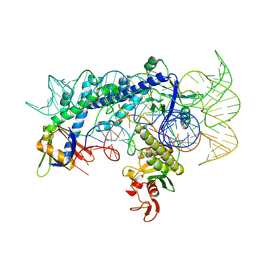 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
6SK0
 
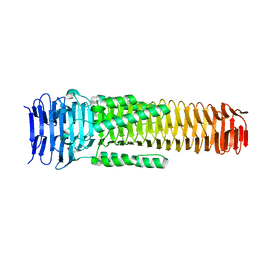 | | The VgrG spike from the Type 6 secretion system | | Descriptor: | Putative type VI secretion protein | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2019-08-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for loading and inhibition of a bacterial T6SS phospholipase effector by the VgrG spike.
Embo J., 39, 2020
|
|
6B5H
 
 | |
1NZ8
 
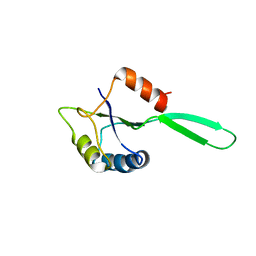 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
7V00
 
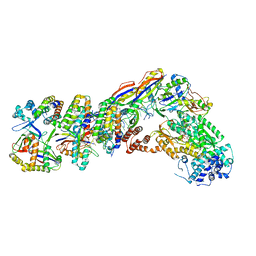 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7UNK
 
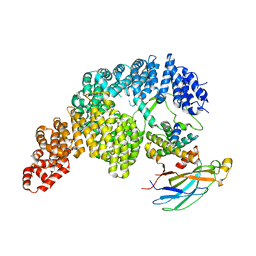 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2GTU
 
 | | LIGAND-FREE HUMAN GLUTATHIONE S-TRANSFERASE M2-2 (E.C.2.5.1.18), MONOCLINIC CRYSTAL FORM | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Patskovska, L.N, Fedorov, A.A, Patskovsky, Y.V, Almo, S.C, Listowsky, I. | | Deposit date: | 1998-05-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The enhanced affinity for thiolate anion and activation of enzyme-bound glutathione is governed by an arginine residue of human Mu class glutathione S-transferases.
J.Biol.Chem., 275, 2000
|
|
2GSA
 
 | |
1NI8
 
 | | H-NS dimerization motif | | Descriptor: | DNA-binding protein H-NS | | Authors: | Bloch, V, Yang, Y, Margeat, E, Chavanieu, A, Aug, M.T, Robert, B, Arold, S, Rimsky, S, Kochoyan, M. | | Deposit date: | 2002-12-22 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The H-NS dimerisation domain defines a new fold contributing to DNA recognition
Nat.Struct.Biol., 10, 2003
|
|
