6NQF
 
 | |
6GP6
 
 | | MamM CTD - Copper form | | Descriptor: | BETA-MERCAPTOETHANOL, COPPER (II) ION, Magnetosome protein MamM | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | MamM CTD - Copper form
To Be Published
|
|
7D2L
 
 | | Crystal structure of the Cas12i1 R-loop complex before target DNA cleavage | | Descriptor: | 12i1-D647A, CITRIC ACID, DNA (26-MER), ... | | Authors: | Zhang, B, Luo, D.Y, Li, Y, OuYang, S.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1.
Nat Commun, 12, 2021
|
|
5CQ4
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-hydroxyphenyl)ethanone, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 3'-Hydroxyacetophenone (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6YRQ
 
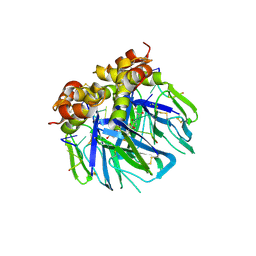 | | Crystal structure of the tetramerization domain of the glycoprotein Gn (Andes virus) at pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-04-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
5LBM
 
 | | The asymmetric tetrameric structure of the formaldehyde sensing transcriptional repressor FrmR from Escherichia coli | | Descriptor: | FORMYL GROUP, Transcriptional repressor FrmR | | Authors: | Bisson, C, Baker, P.J, Green, J, Chivers, P.T. | | Deposit date: | 2016-06-16 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The mechanism of a formaldehyde-sensing transcriptional regulator.
Sci Rep, 6, 2016
|
|
5IEH
 
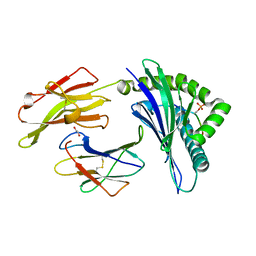 | |
6Q12
 
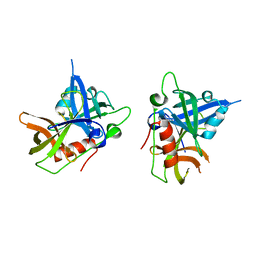 | | Structure of pro-Esp mutant- S66V | | Descriptor: | Glutamyl endopeptidase | | Authors: | Manne, K, Narayana, S.V.L. | | Deposit date: | 2019-08-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the role of the N-terminus in the activation and function of extracellular serine protease from Staphylococcus epidermidis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6GHZ
 
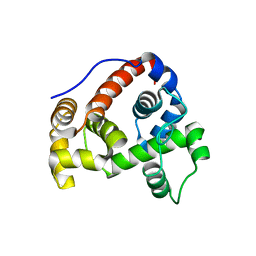 | |
5LBG
 
 | | Crystal structure of the Mycobacterium tuberculosis L,D-transpeptidase-2 (LdtMt2) BC-module with faropenem-derived adduct at the active site cysteine-354 | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, L,D-transpeptidase 2 | | Authors: | Steiner, E.M, Schnell, R, Schneider, G. | | Deposit date: | 2016-06-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Binding and processing of beta-lactam antibiotics by the transpeptidase LdtMt2 from Mycobacterium tuberculosis.
FEBS J., 284, 2017
|
|
8OLH
 
 | |
6GI6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
7D8C
 
 | | Crystal structure of the Cas12i1-crRNA binary complex | | Descriptor: | 12i1, CITRIC ACID, RNA (3-MER), ... | | Authors: | Zhang, B, Luo, D.Y, Li, Y, OuYang, S.Y. | | Deposit date: | 2020-10-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1.
Nat Commun, 12, 2021
|
|
5D7G
 
 | |
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5D80
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | Authors: | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | Deposit date: | 2015-08-14 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (6.202 Å) | | Cite: | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
8P26
 
 | | Crystal structure of Arabidopsis thaliana PAXX | | Descriptor: | U2 small nuclear ribonucleoprotein auxiliary factor-like protein | | Authors: | Ochi, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plant PAXX has an XLF-like function and stimulates DNA end joining by the Ku-DNA ligase IV/XRCC4 complex.
Plant J., 116, 2023
|
|
5KZD
 
 | | N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus with bound sialic acid alditol | | Descriptor: | (2~{S},4~{S},5~{R},6~{R},7~{S},8~{R})-5-acetamido-2,4,6,7,8,9-hexakis(oxidanyl)nonanoic acid, N-acetylneuraminate lyase | | Authors: | North, R.A, Watson, A.J.A, Pearce, F.G, Muscroft-Taylor, A.C, Friemann, R, Fairbanks, A.J, Dobson, R.C.J. | | Deposit date: | 2016-07-25 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structure and inhibition of N-acetylneuraminate lyase from methicillin-resistant Staphylococcus aureus.
FEBS Lett., 590, 2016
|
|
7D3J
 
 | | Crystal structure of the Cas12i1 R-loop complex after target DNA cleavage | | Descriptor: | 12i1-WT, CITRIC ACID, DNA (23-MER), ... | | Authors: | Zhang, B, Luo, D.Y, Li, Y, OuYang, S.Y. | | Deposit date: | 2020-09-19 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1.
Nat Commun, 12, 2021
|
|
6GPJ
 
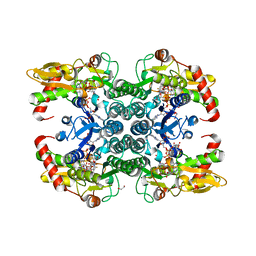 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6GQA
 
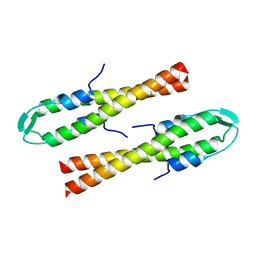 | | Cell division regulator S. pneumoniae GpsB | | Descriptor: | Cell cycle protein GpsB | | Authors: | Lewis, R.J, Rutter, Z.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The cell cycle regulator GpsB functions as cytosolic adaptor for multiple cell wall enzymes.
Nat Commun, 10, 2019
|
|
5W1I
 
 | |
6TVL
 
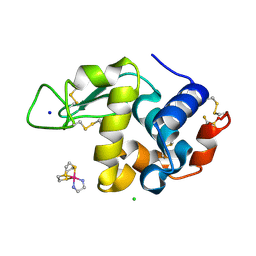 | | Hen Egg White Lysozyme in complex with a "half sandwich"-type Ru(II) coordination compound | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | High-resolution crystal structures of a "half sandwich"-type Ru(II) coordination compound bound to hen egg-white lysozyme and proteinase K.
J.Biol.Inorg.Chem., 25, 2020
|
|
6GQI
 
 | |
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
