6WW9
 
 | | Crystal structure of human REV7(R124A)-SHLD3(35-58) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, Shieldin complex subunit 3 | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L4S
 
 | |
2KV2
 
 | |
7XYB
 
 | | The cryo-EM structure of an AlpA-loaded complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
6A2T
 
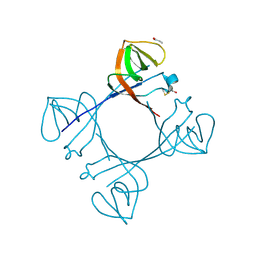 | | Mycobacterium tuberculosis LexA C-domain K197A | | Descriptor: | ACRYLIC ACID, LexA repressor | | Authors: | Chandran, A.V, Srikalaivani, R, Paul, A, Vijayan, M. | | Deposit date: | 2018-06-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical characterization of Mycobacterium tuberculosis LexA and structural studies of its C-terminal segment.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6RNX
 
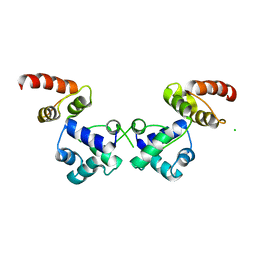 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6VEN
 
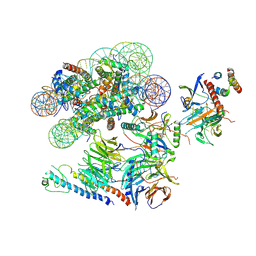 | |
8DK9
 
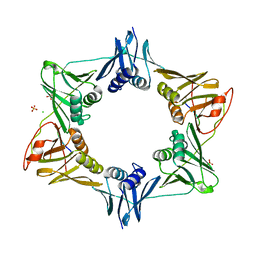 | | Sliding-clamp-DinX peptide | | Descriptor: | ACETYL GROUP, AMINO GROUP, Beta sliding clamp, ... | | Authors: | Kapur, M.K, Gray, O.J, Honzatko, R.H, Nelson, S.N. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of sliding clamp with mycobacterial polymerases
To Be Published
|
|
3KZ7
 
 | |
6SDG
 
 | |
4A0Z
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
2C9O
 
 | | 3D Structure of the human RuvB-like helicase RuvBL1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RUVB-LIKE 1 | | Authors: | Matias, P.M, Gorynia, S, Donner, P, Carrondo, M.A. | | Deposit date: | 2005-12-14 | | Release date: | 2006-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human AAA+ protein RuvBL1.
J. Biol. Chem., 281, 2006
|
|
4A0Y
 
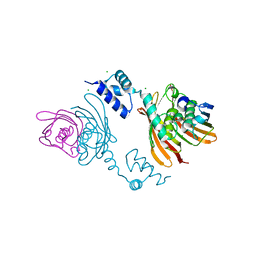 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4YN8
 
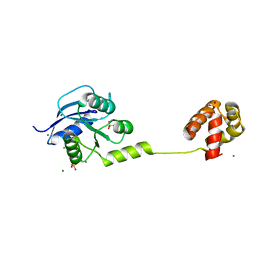 | | Crystal Structure of Response Regulator ChrA in Heme-Sensing Two Component System | | Descriptor: | MAGNESIUM ION, Response regulator ChrA, SULFATE ION | | Authors: | Doi, A, Nakamura, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2015-03-09 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the response regulator ChrA in the haem-sensing two-component system of Corynebacterium diphtheriae.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
3MEF
 
 | |
3IKV
 
 | |
2W48
 
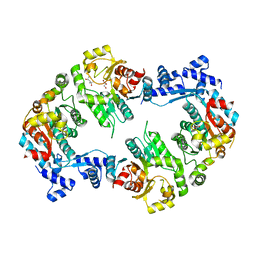 | | Crystal structure of the Full-length Sorbitol Operon Regulator SorC from Klebsiella pneumoniae | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | de Sanctis, D, McVey, C.E, Enguita, F.J, Carrondo, M.A. | | Deposit date: | 2008-11-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Full-Length Sorbitol Operon Regulator Sorc from Klebsiella Pneumoniae: Structural Evidence for a Novel Transcriptional Regulation Mechanism.
J.Mol.Biol., 387, 2009
|
|
2N5G
 
 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
5HS9
 
 | |
6NON
 
 | | Structure of Cyanthece apo McdA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cobyrinic acid ac-diamide synthase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
6NOP
 
 | | Structure of Cyanothece McdA(D38A)-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobyrinic acid ac-diamide synthase, MAGNESIUM ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
2O20
 
 | | Crystal structure of transcription regulator CcpA of Lactococcus lactis | | Descriptor: | CHLORIDE ION, Catabolite control protein A, SULFATE ION | | Authors: | Loll, B, Kowalczyk, M, Alings, C, Chieduch, A, Bardowski, J, Saenger, W, Biesiadka, J. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the transcription regulator CcpA from Lactococcus lactis
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1A04
 
 | | THE STRUCTURE OF THE NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL IN THE MONOCLINIC C2 CRYSTAL FORM | | Descriptor: | NITRATE/NITRITE RESPONSE REGULATOR PROTEIN NARL | | Authors: | Baikalov, I, Schroder, I, Kaczor-Grzeskowiak, M, Cascio, D, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 1997-12-08 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NarL dimerization? Suggestive evidence from a new crystal form
Biochemistry, 37, 1998
|
|
5HS7
 
 | |
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | Authors: | Xie, W, Patel, D.J. | | Deposit date: | 2021-01-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
