6OH1
 
 | | IgA1 Protease G5 domain structure | | Descriptor: | Immunoglobulin A1 protease | | Authors: | Eisenmesser, E.Z, Chi, Y.C, Paukovich, N, Redzic, J.S, Rahkola, J.T, Janoff, E.N. | | Deposit date: | 2019-04-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Streptococcus pneumoniae G5 domains bind different ligands.
Protein Sci., 28, 2019
|
|
1GA3
 
 | |
5HTD
 
 | | Recombinant bovine beta-lactoglobulin variant L1A/I2S with endogenous ligand (sBlgB#1) | | Descriptor: | Beta-lactoglobulin, MYRISTIC ACID | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-01-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
5HTE
 
 | | Recombinant bovine beta-lactoglobulin variant L1A/I2S (sBlgB#2) | | Descriptor: | Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-01-26 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
7RYE
 
 | |
6HMP
 
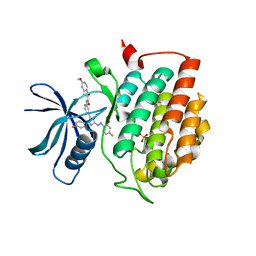 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azoimidazole-based Inhibitor (compound 3) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[5-(4-fluorophenyl)-2-[(~{E})-(4-fluorophenyl)diazenyl]-3-methyl-imidazol-4-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
6HMR
 
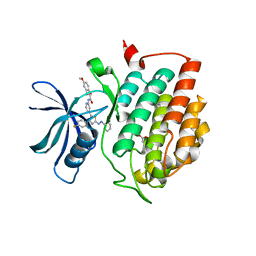 | | Crystal structure of human Casein Kinase I delta in complex with a photoswitchable 2-Azothiazole-based inhibitor (compound 2) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-[(~{E})-phenyldiazenyl]-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Casein kinase I isoform delta, MALONIC ACID | | Authors: | Pichlo, C, Schehr, M, Charl, J, Brunstein, E, Peifer, C, Baumann, U. | | Deposit date: | 2018-09-12 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
3KXP
 
 | | Crystal Structure of E-2-(Acetamidomethylene)succinate Hydrolase | | Descriptor: | Alpha-(N-acetylaminomethylene)succinic acid hydrolase, CHLORIDE ION | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure determination and characterization of the vitamin B(6) degradative enzyme (E)-2-(acetamidomethylene)succinate hydrolase.
Biochemistry, 49, 2010
|
|
1LTB
 
 | |
7P49
 
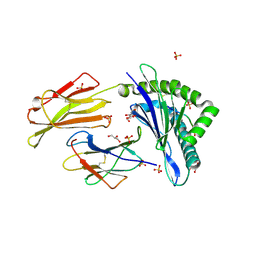 | | HLA-E*01:03 in complex with Mtb14 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
7P4B
 
 | | HLA-E*01:03 in complex with IL9 | | Descriptor: | Beta-2-microglobulin, ESAT-6-like protein EsxH, GLYCEROL, ... | | Authors: | Walters, L.C, Gillespie, G.M. | | Deposit date: | 2021-07-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Cell Rep, 39, 2022
|
|
1MBB
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-3(N-ACETYLGLUCOSAMINYL)BUTYRIC ACID | | Authors: | Benson, T.E, Lees, W.J, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (E)-enolbutyryl-UDP-N-acetylglucosamine as a mechanistic probe of UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 35, 1996
|
|
4HR7
 
 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
9AZS
 
 | |
8CXH
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C10 heavy chain, C10 light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXG
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | Ankyrin repeat family A protein 2,Envelope E protein, C8 scFv heavy chain, C8 scFv light chain, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
8CXI
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
7UWH
 
 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate | | Descriptor: | DNA (59-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
7UWE
 
 | | CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Hao, Z.T, Grower, M, Bharati, B, Proshkin, S, Epshtein, V, Svetlov, V, Nudler, E, Shamovsky, I. | | Deposit date: | 2022-05-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | RNA polymerase drives ribonucleotide excision DNA repair in E. coli.
Cell, 186, 2023
|
|
8UIZ
 
 | | In situ human P-E state 80S ribosome | | Descriptor: | 18S rRNA [Homo sapiens], 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wei, Z, Yong, X. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | In situ human P-E state 80S ribosome
To Be Published
|
|
4C16
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
7QB2
 
 | | Pim1 in complex with (E)-4-((6-amino-1-methyl-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(E)-(6-azanyl-1-methyl-2-oxidanylidene-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Pimtide, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
8ELM
 
 | | Apo human biliverdin reductase beta (293K) | | Descriptor: | Flavin reductase (NADPH), SODIUM ION | | Authors: | McLeod, M.J, Eisenmesser, E.Z, Lee, E, Thorne, R.E. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identifying structural and dynamic changes during the Biliverdin Reductase B catalytic cycle.
Front Mol Biosci, 10, 2023
|
|
4U7E
 
 | | The crystal structure of the complex of LIP5 NTD and IST1 MIM | | Descriptor: | IST1 homolog, Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Guo, E.Z, Xu, Z. | | Deposit date: | 2014-07-30 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinct Mechanisms of Recognizing Endosomal Sorting Complex Required for Transport III (ESCRT-III) Protein IST1 by Different Microtubule Interacting and Trafficking (MIT) Domains.
J.Biol.Chem., 290, 2015
|
|
6OPL
 
 | |
