8G0A
 
 | |
8FL8
 
 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3ZO6
 
 | |
2W5J
 
 | | Structure of the c14-rotor ring of the proton translocating chloroplast ATP synthase | | Descriptor: | ATP SYNTHASE C CHAIN, CHLOROPLASTIC | | Authors: | Vollmar, M, Schlieper, D, Winn, M, Buechner, C, Groth, G. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase.
J. Biol. Chem., 284, 2009
|
|
1L6T
 
 | |
2X2V
 
 | | Structural basis of a novel proton-coordination type in an F1Fo-ATP synthase rotor ring | | Descriptor: | ATP SYNTHASE SUBUNIT C, SODIUM ION, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Preiss, L, Yildiz, O, Hicks, D.B, Krulwich, T.A, Meier, T. | | Deposit date: | 2010-01-18 | | Release date: | 2010-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A New Type of Proton Coordination in an F(1)F(O)- ATP Synthase Rotor Ring.
Plos Biol., 8, 2010
|
|
2KHK
 
 | | NMR solution structure of the b30-82 domain of subunit b of Escherichia coli F1FO ATP synthase | | Descriptor: | ATP synthase subunit b | | Authors: | Priya, R, Biukovic, G, Gayen, S, Vivekanandan, S, Gruber, G. | | Deposit date: | 2009-04-08 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure, determined by nuclear magnetic resonance, of the b30-82 domain of subunit b of Escherichia coli F1Fo ATP synthase
J.Bacteriol., 191, 2009
|
|
4CBK
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, SODIUM ION, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
4CBJ
 
 | | The c-ring ion binding site of the ATP synthase from Bacillus pseudofirmus OF4 is adapted to alkaliphilic cell physiology | | Descriptor: | ATP SYNTHASE SUBUNIT C, DODECYL-BETA-D-MALTOSIDE, TRIS(HYDROXYETHYL)AMINOMETHANE, ... | | Authors: | Preiss, L, Yildiz, O, Meier, T. | | Deposit date: | 2013-10-14 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The C-Ring Ion-Binding Site of the ATP Synthase from Bacillus Pseudofirmus of4 is Adapted to Alkaliphilic Lifestyle.
Mol.Microbiol., 92, 2014
|
|
7Y5C
 
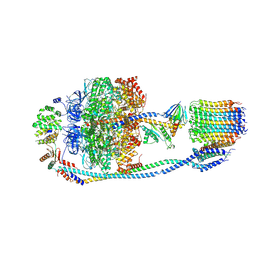 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5B
 
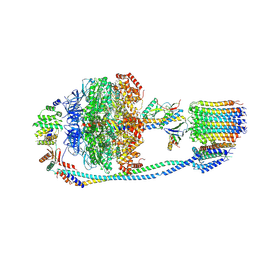 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
6J54
 
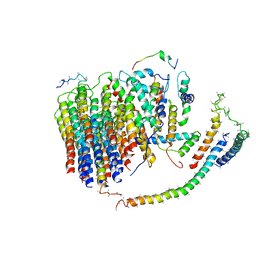 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J5A
 
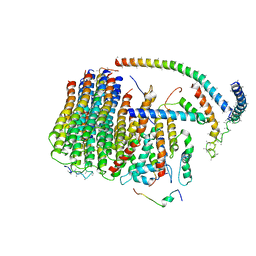 | | Cryo-EM structure of the mammalian DP-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
5FIJ
 
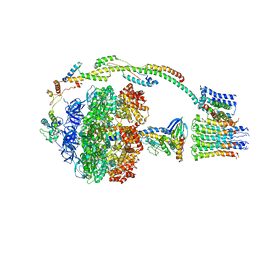 | | Bovine mitochondrial ATP synthase state 2c | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIK
 
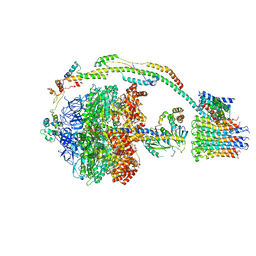 | | Bovine mitochondrial ATP synthase state 3a | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
5FIL
 
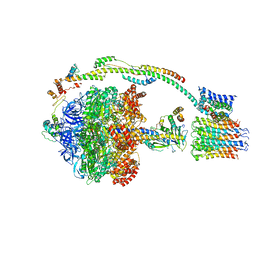 | | Bovine mitochondrial ATP synthase state 3b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
6TQJ
 
 | | Crystal structure of the c14 ring of the F1FO ATP synthase from spinach chloroplast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ATP synthase subunit c, chloroplastic, ... | | Authors: | Kovalev, K, Gushchin, I, Vlasov, A, Round, E, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual features of the c-ring of F1FOATP synthases.
Sci Rep, 9, 2019
|
|
1C99
 
 | |
7NL9
 
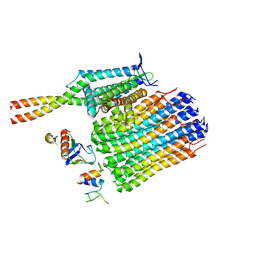 | | Mycobacterium smegmatis ATP synthase Fo state 3 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKP
 
 | | Mycobacterium smegmatis ATP synthase Fo state 2 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NK9
 
 | | Mycobacterium smegmatis ATP synthase Fo domain state 1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-11-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKK
 
 | | Mycobacterium smegmatis ATP synthase rotor state 2 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKB
 
 | | Mycobacterium smegmatis ATP synthase rotor state 1 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-17 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NKN
 
 | | Mycobacterium smegmatis ATP synthase rotor state 3 | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Montgomery, M.G, Petri, J, Spikes, T.E, Walker, J.E. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structure of the ATP synthase from Mycobacterium smegmatis provides targets for treating tuberculosis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1ATY
 
 | |
