3S3Z
 
 | |
3RY9
 
 | |
3RZ1
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, N-(2,2,3,3,4,4,5,5,5-nonafluoropentyl)-4-sulfamoylbenzamide, ... | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
3RU3
 
 | | Crystal structure of tm0922, a fusion of a domain of unknown function and ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Thermotoga maritima soaked with NADPH and ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-6-HYDROXY-1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Lesley, S.A, Minor, W. | | Deposit date: | 2011-05-04 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RUL
 
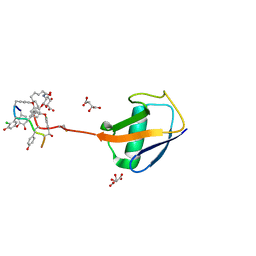 | | New strategy to analyze structures of glycopeptide-target complexes | | Descriptor: | 10-METHYLUNDECANOIC ACID, 2-amino-2-deoxy-beta-D-glucopyranuronic acid, CHLORIDE ION, ... | | Authors: | Economou, N.J, Nahoum, V, Weeks, S.D, Grasty, K.C, Loll, P.J. | | Deposit date: | 2011-05-05 | | Release date: | 2012-06-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A carrier protein strategy yields the structure of dalbavancin.
J.Am.Chem.Soc., 134, 2012
|
|
3RZD
 
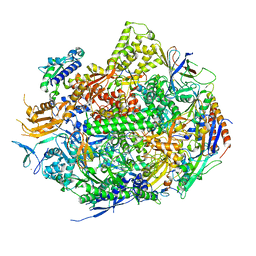 | | RNA Polymerase II Initiation Complex with a 5-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3RUP
 
 | |
3RW6
 
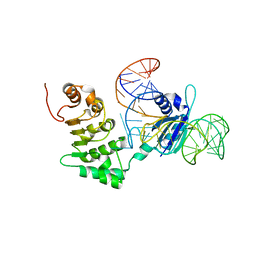 | | Structure of nuclear RNA export factor TAP bound to CTE RNA | | Descriptor: | Nuclear RNA export factor 1, constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | | Authors: | Teplova, M, Khin, N.W, Patel, D.J, Izaurralde, E. | | Deposit date: | 2011-05-07 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function studies of nucleocytoplasmic transport of retroviral genomic RNA by mRNA export factor TAP.
Nat.Struct.Mol.Biol., 18, 2011
|
|
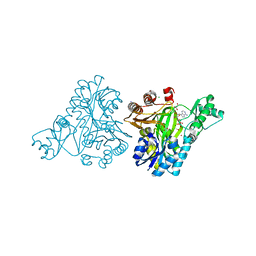
3RV4
 
 | |
3S06
 
 | |
3RUY
 
 | | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis | | Descriptor: | Ornithine aminotransferase | | Authors: | Anderson, S.M, Wawrzak, Z, Brunzelle, J.S, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-05 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Ornithine-oxo acid transaminase RocD from Bacillus anthracis
To be Published
|
|
3RZO
 
 | | RNA Polymerase II Initiation Complex with a 4-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-12 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3RVF
 
 | | FXR with SRC1 and GSK2034 | | Descriptor: | 5-(4-{[3-(2,6-dichlorophenyl)-5-(propan-2-yl)-1,2-oxazol-4-yl]methoxy}phenyl)-1H-indole-2-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2011-05-06 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformationally constrained farnesoid X receptor (FXR) agonists: Alternative replacements of the stilbene.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3RWX
 
 | |
3S1R
 
 | | RNA Polymerase II Initiation Complex with a 5-nt 3'-deoxy RNA soaked with GTP | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3S2I
 
 | | Crystal Structure of FurX NADH+:Furfuryl alcohol II | | Descriptor: | FURFURAL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Hayes, R, Sanchez, E.J, Webb, B.N, Hooper, T, Nissen, M.S, Li, Q, Xun, L. | | Deposit date: | 2011-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and furfural reduction mechanism of a bacterial zinc-dependent alcohol dehydrogenase
To be Published
|
|
3S0H
 
 | |
3S16
 
 | | RNA Polymerase II Initiation Complex with an 8-nt RNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*GP*AP*TP*AP*AP*GP*CP*AP*GP*AP*CP*GP*AP*TP*CP*CP*TP*CP*TP*CP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Liu, X, Bushnell, D.A, Silva, D.A, Huang, X, Kornberg, R.D. | | Deposit date: | 2011-05-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.241 Å) | | Cite: | Initiation complex structure and promoter proofreading.
Science, 333, 2011
|
|
3RV3
 
 | |
3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
3QH7
 
 | | 2.5 A resolution structure of Se-Met labeled CT296 from Chlamydia trachomatis | | Descriptor: | CT296 | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J. Bacteriol., 193, 2011
|
|
3QHX
 
 | |
3QNK
 
 | |
3QIX
 
 | | Crystal Structure of BoNT/A LC with Zinc bound | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ZINC ION | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QK7
 
 | | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001 | | Descriptor: | Transcriptional regulators | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001
To be Published
|
|
