8SLB
 
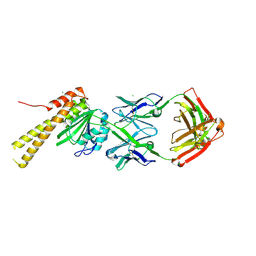 | | X-ray structure of CorA N-terminal domain in complex with conformation-specific synthetic antibody C12 | | Descriptor: | CHLORIDE ION, Cobalt/magnesium transport protein CorA, sAB C12 Heavy Chain, ... | | Authors: | Dominik, P.K, Erramilli, S.K, Reddy, B.G, Kossiakoff, A.A. | | Deposit date: | 2023-04-21 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA.
J.Mol.Biol., 435, 2023
|
|
3CGG
 
 | |
3CLR
 
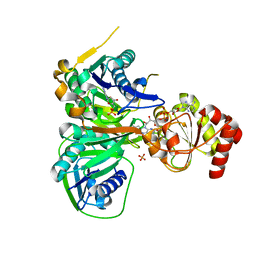 | | Crystal structure of the R236A ETF mutant from M. methylotrophus | | Descriptor: | ADENOSINE MONOPHOSPHATE, Electron transfer flavoprotein subunit alpha, Electron transfer flavoprotein subunit beta, ... | | Authors: | Katona, G, Leys, D. | | Deposit date: | 2008-03-20 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Biochemistry, 47, 2008
|
|
7S2E
 
 | |
3CGX
 
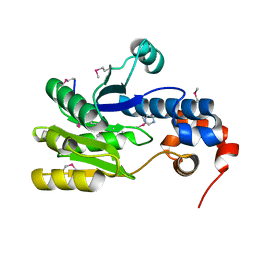 | |
7S2Q
 
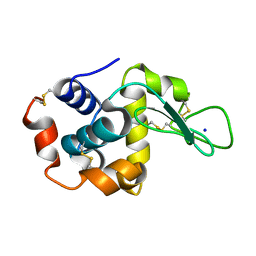 | |
3CH5
 
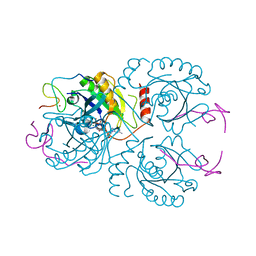 | | The crystal structure of the RanGDP-Nup153ZnF2 complex | | Descriptor: | Fragment of Nuclear pore complex protein Nup153, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vetter, I.R, Schrader, N. | | Deposit date: | 2008-03-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex
Structure, 16, 2008
|
|
3CHJ
 
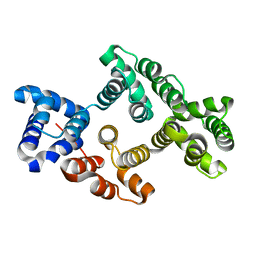 | | Crystal Structure of Alpha-14 Giardin | | Descriptor: | Alpha-14 giardin, CALCIUM ION | | Authors: | Pathuri, P, Luecke, H. | | Deposit date: | 2008-03-10 | | Release date: | 2009-01-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Apo and calcium-bound crystal structures of cytoskeletal protein alpha-14 giardin (annexin E1) from the intestinal protozoan parasite Giardia lamblia
J.Mol.Biol., 385, 2009
|
|
7S31
 
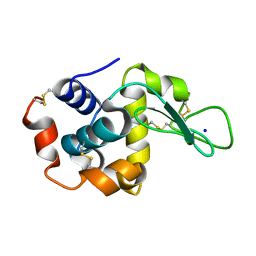 | |
3CHM
 
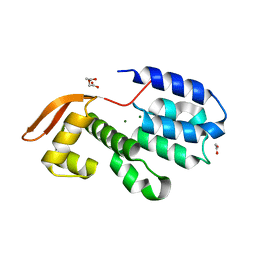 | |
6ISH
 
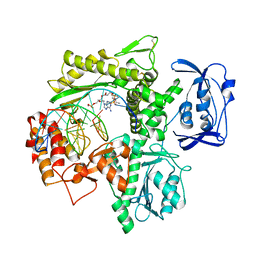 | | Structure of 9N-I DNA polymerase incorporation with 3'-AL in the active site | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*CP*TP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*CP*(DAL))-3'), ... | | Authors: | Linwu, S.W, Maestre-Reyna, M, Tsai, M.D, Tu, Y.H, Chang, W.H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermococcus sp. 9°N DNA polymerase exhibits 3'-esterase activity that can be harnessed for DNA sequencing.
Commun Biol, 2, 2019
|
|
6IU8
 
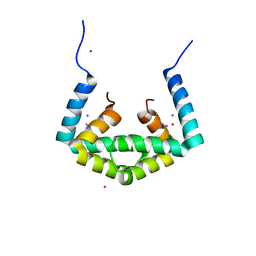 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
3CQT
 
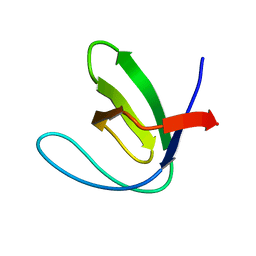 | | N53I V55L MUTANT of FYN SH3 DOMAIN | | Descriptor: | Proto-oncogene tyrosine-protein kinase Fyn | | Authors: | Neculai, A.M, Zarrine-Afsar, A, Howell, P.L, Davidson, A, Chan, H.S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Theoretical and experimental demonstration of the importance of specific nonnative interactions in protein folding.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8SXO
 
 | |
3CQY
 
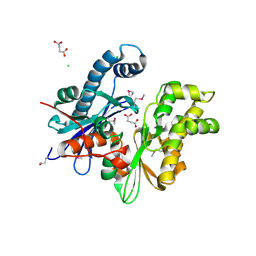 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
6DYY
 
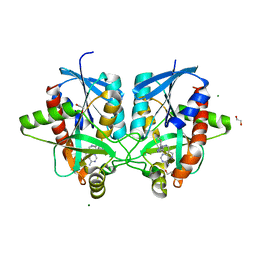 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
6IVK
 
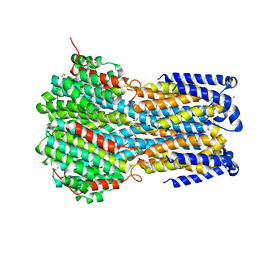 | | Crystal structure of a membrane protein G175A | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kittredge, A, Fukuda, F, Zhang, Y, Yang, T. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Dual Ca2+-dependent gates in human Bestrophin1 underlie disease-causing mechanisms of gain-of-function mutations.
Commun Biol, 2, 2019
|
|
3CS2
 
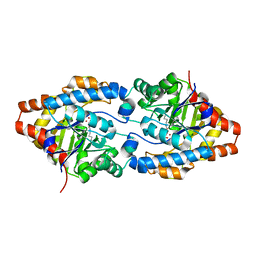 | | Crystal structure of PTE G60A mutant | | Descriptor: | CACODYLATE ION, COBALT (II) ION, Parathion hydrolase | | Authors: | Kim, J, Almo, S.C. | | Deposit date: | 2008-04-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
3CSW
 
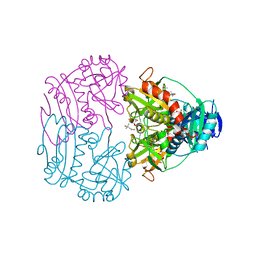 | |
5K95
 
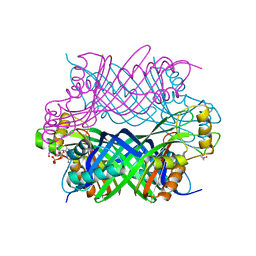 | |
3CUO
 
 | | Crystal structure of the predicted DNA-binding transcriptional regulator from E. coli | | Descriptor: | Uncharacterized HTH-type transcriptional regulator ygaV | | Authors: | Zhang, R, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the predicted DNA-binding transcriptional regulator from E. coli.
To be Published
|
|
3CUX
 
 | |
6IG6
 
 | |
3CUV
 
 | | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyritsi, C, Chrysina, E.D, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G. | | Deposit date: | 2008-04-17 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Tracking structure activity relationships of glycogen phosphorylase inhibitors: synthesis, kinetic and crystallographic evaluation of analogues of N-(-D-glucopyranosyl)-N'-oxamides
To be Published
|
|
5K9G
 
 | | Crystal Structure of GTP Cyclohydrolase-IB with Tris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, ... | | Authors: | Alvarez, J, Stec, B, Swairjo, M.A. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism and catalytic strategy of the prokaryotic-specific GTP cyclohydrolase-IB.
Biochem.J., 474, 2017
|
|
