2P31
 
 | | Crystal structure of human glutathione peroxidase 7 | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 7 | | Authors: | Kavanagh, K.L, Johansson, C, Papagrigoriou, E, Kochan, G, Umeano, C, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-08 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human glutathione peroxidase 7
To be Published
|
|
6O7X
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4AZK
 
 | | Structural basis of L-phosphoserine binding to Bacillus alcalophilus phosphoserine aminotransferase | | Descriptor: | CHLORIDE ION, PHOSPHOSERINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Battula, P, Dubnovitsky, A.P, Papageorgiou, A.C. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Structural Basis of L-Phosphoserine Binding to Bacillus Alcalophilus Phosphoserine Aminotransferase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6TCL
 
 | | Photosystem I tetramer | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
6TAA
 
 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF ASPERGILLUS ORYZAE (TAKA) ALPHA-AMYLASE: AN APPLICATION OF THE SIMULATED-ANNEALING METHOD | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Swift, H.J, Brady, L, Derewenda, Z.S, Dodson, E.J, Turkenburg, J.P, Wilkinson, A.J. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and molecular model refinement of Aspergillus oryzae (TAKA) alpha-amylase: an application of the simulated-annealing method.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
4B1X
 
 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
3CQI
 
 | |
6OH4
 
 | |
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
3CTT
 
 | | Crystal complex of N-terminal Human Maltase-Glucoamylase with Casuarine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CASUARINE, GLYCEROL, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2008-04-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Total syntheses of casuarine and its 6-O-alpha-glucoside: complementary inhibition towards glycoside hydrolases of the GH31 and GH37 families
Chemistry, 15, 2009
|
|
6TZV
 
 | | Crystal Structure of the carboxyltransferase subunit of ACC (AccD6) in complex with inhibitor Phenyl-Cyclodiaone from Mycobacterium tuberculosis | | Descriptor: | 4'-[(6-chloro-1,3-benzothiazol-2-yl)oxy]-6-hydroxy-4,4-dimethyl-4,5-dihydro[1,1'-biphenyl]-2(3H)-one, Probable propionyl-CoA carboxylase beta chain 6 | | Authors: | Reddy, M.C.M, Zhou, N, Sacchettini, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-08-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.388 Å) | | Cite: | Novel herbicidal derivatives that inhibit carboxyltransferase subunit of the acetyl-CoA carboxylase in Mycobacterium tuberculosis
To Be Published
|
|
6U34
 
 | | Binary complex of native hAChE with oxime reactivator RS194B | | Descriptor: | (2E)-N-[2-(azepan-1-yl)ethyl]-2-(hydroxyimino)acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
4CPF
 
 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
2P0W
 
 | | Human histone acetyltransferase 1 (HAT1) | | Descriptor: | ACETAMIDE, ACETATE ION, ACETYL COENZYME *A, ... | | Authors: | Wu, H, Min, J, Zeng, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-01 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human histone acetyltransferase 1 (HAT1) in complex with acetylcoenzyme A and histone peptide H4
To be Published
|
|
2P1D
 
 | |
6OEQ
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3CVS
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
6OH2
 
 | |
6U3E
 
 | |
2P5W
 
 | | Crystal structures of high affinity human T-cell receptors bound to pMHC reveal native diagonal binding geometry | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Sami, M, Rizkallah, P.J, Dunn, S, Li, Y, Moysey, R, Vuidepot, A, Baston, E, Todorov, P, Molloy, P, Gao, F, Boulter, J.M, Jakobsen, B.K. | | Deposit date: | 2007-03-16 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of high affinity human T-cell receptors bound to peptide major
histocompatibility complex reveal native diagonal binding geometry
Protein Eng.Des.Sel., 20, 2007
|
|
6OQV
 
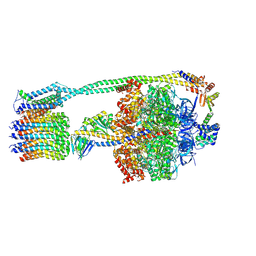 | | E. coli ATP Synthase State 2b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
4D3J
 
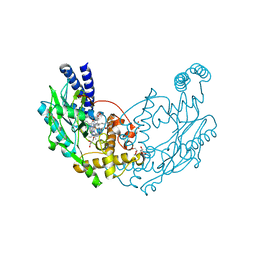 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
3FQD
 
 | | Crystal Structure of the S. pombe Rat1-Rai1 Complex | | Descriptor: | 5'-3' exoribonuclease 2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
6TZC
 
 | | Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin | | Descriptor: | Apoptosis regulator Bcl-2 homolog, Beclin-1, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Banjara, S, Kvansakul, M, Hinds, M.G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of African Swine Fever Virus A179L with the Autophagy Regulator Beclin.
Viruses, 11, 2019
|
|
3FTO
 
 | | Crystal structure of OppA in a open conformation | | Descriptor: | Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Oktaviani, N.A, Fusetti, F, Thunnissen, A.-M.W.H, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2009-01-13 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Selenomethionine incorporation in proteins expressed in Lactococcus lactis.
Protein Sci., 18, 2009
|
|
