4BKG
 
 | | crystal structure of human diSUMO-2 | | Descriptor: | SMALL UBIQUITIN-RELATED MODIFIER 2 | | Authors: | Keusekotten, K, Bade, V.N, Meyer-Teschendorf, K, Sriramachandran, A, Fischer-Schrader, K, Krause, A, Horst, C, Hofmann, K, Dohmen, R.J, Praefcke, G.J.K. | | Deposit date: | 2013-04-25 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Multivalent Interactions of the Sumo-Interaction Motifs in the Ring-Finger Protein 4 (Rnf4) Determine the Specificity for Chains of the Small Ubiquitin-Related Modifier (Sumo).
Biochem.J., 457, 2014
|
|
1RQ2
 
 | | MYCOBACTERIUM TUBERCULOSIS FTSZ IN COMPLEX WITH CITRATE | | Descriptor: | CITRIC ACID, Cell division protein ftsZ | | Authors: | Leung, A.K.W, White, E.L, Ross, L.J, Reynolds, R.C, DeVito, J.A, Borhani, D.W. | | Deposit date: | 2003-12-04 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches.
J.Mol.Biol., 342, 2004
|
|
5MD7
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=-12 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
7A1I
 
 | | Crystal structure of the BILBO2/FPC4 complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BILBO1_N domain-containing protein, ... | | Authors: | Dong, G. | | Deposit date: | 2020-08-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and functional studies of the first tripartite protein complex at the Trypanosoma brucei flagellar pocket collar.
Plos Pathog., 17, 2021
|
|
3MT6
 
 | | Structure of ClpP from Escherichia coli in complex with ADEP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACYLDEPSIPEPTIDE 1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Chung, Y.S. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP.
Chem.Biol., 17, 2010
|
|
4B95
 
 | | pVHL-EloB-EloB-EloC complex_(2S,4R)-1-(2-chlorophenyl)carbonyl-N-[(4-chlorophenyl)methyl]-4-oxidanyl-pyrrolidine-2-carboxamide bound | | Descriptor: | (2S,4R)-1-(2-chlorophenyl)carbonyl-N-[(4-chlorophenyl)methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Buckley, D.L, Gustafson, J.L, VanMolle, I, Roth, A.G, SeopTae, H, Gareiss, P.C, Jorgensen, W.L, Ciulli, A, Crews, C.M. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1Alpha
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4BJS
 
 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
7AD8
 
 | | Core TFIIH-XPA-DNA complex with modelled p62 subunit | | Descriptor: | DNA (49-MER), DNA repair protein complementing XP-A cells, General transcription and DNA repair factor IIH helicase subunit XPB, ... | | Authors: | Koelmel, W, Kuper, J, Kisker, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The TFIIH subunits p44/p62 act as a damage sensor during nucleotide excision repair.
Nucleic Acids Res., 48, 2020
|
|
4GXP
 
 | | Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA | | Descriptor: | Beta-glucosidase Chimeric protein | | Authors: | Smith, M.A, Romero, P.A, Wu, T, Brustad, E.M, Arnold, F.H. | | Deposit date: | 2012-09-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chimeragenesis of distantly-related proteins by noncontiguous recombination.
Protein Sci., 22, 2013
|
|
4BK8
 
 | | Superoxide reductase (Neelaredoxin) from Ignicoccus hospitalis | | Descriptor: | DESULFOFERRODOXIN, FERROUS IRON-BINDING REGION, FE (III) ION | | Authors: | Romao, C.V, Matias, P.M, Pinho, F.G, Sousa, C.M, Barradas, A.R, Pinto, A.F, Teixeira, M, Bandeiras, T.M. | | Deposit date: | 2013-04-22 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Structure of a Natural Sor Mutant
To be Published
|
|
3MFU
 
 | | CASK-4M CaM Kinase Domain, AMPPNP-Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peripheral plasma membrane protein CASK | | Authors: | Wahl, M.C, Mukherjee, K. | | Deposit date: | 2010-04-03 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of CASK into a Mg2+-sensitive kinase.
Sci.Signal., 3, 2010
|
|
5MD9
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=6 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
4ARH
 
 | | X ray structure of the periplasmic zinc binding protein ZinT from Salmonella enterica | | Descriptor: | METAL-BINDING PROTEIN YODA, SULFATE ION | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
3N86
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 4 | | Descriptor: | (1R,5R)-1,5-dihydroxy-4-oxo-3-[3-oxo-3-(phenylamino)propyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3MIB
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
4PJ1
 
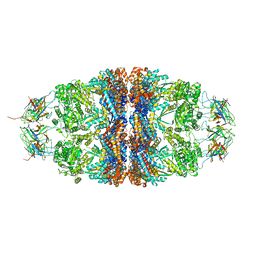 | | Crystal structure of the human mitochondrial chaperonin symmetrical 'football' complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Frolow, F, Azem, A, Nisemblat, S. | | Deposit date: | 2014-05-10 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the human mitochondrial chaperonin symmetrical football complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3MFY
 
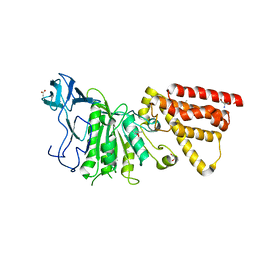 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
1R4M
 
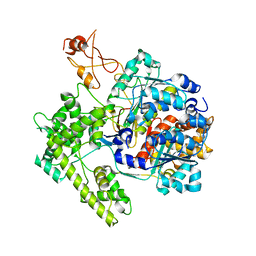 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex | | Descriptor: | Ubiquitin-like protein NEDD8, ZINC ION, amyloid beta precursor protein-binding protein 1, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
1R71
 
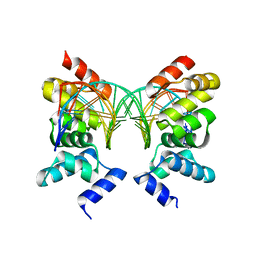 | | Crystal Structure of the DNA binding domain of KorB in complex with the operator DNA | | Descriptor: | 5'-D(*AP*(BRU)P*TP*TP*TP*AP*GP*CP*GP*GP*CP*TP*AP*AP*AP*AP*G)-3', 5'-D(*CP*(BRU)P*TP*TP*TP*AP*GP*CP*CP*GP*CP*TP*AP*AP*AP*AP*(BRU))-3', Transcriptional repressor protein korB | | Authors: | Khare, D, Ziegelin, G, Lanka, E, Heinemann, U. | | Deposit date: | 2003-10-17 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3MPS
 
 | | Peroxide Bound Oxidized Rubrerythrin from Pyrococcus furiosus | | Descriptor: | FE (III) ION, HYDROGEN PEROXIDE, MU-OXO-DIIRON, ... | | Authors: | Dillard, B.D, Adams, M.W.W, Lanzilotta, W.N. | | Deposit date: | 2010-04-27 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A cryo-crystallographic time course for peroxide reduction by rubrerythrin from Pyrococcus furiosus.
J.Biol.Inorg.Chem., 16, 2011
|
|
5MJS
 
 | | S. pombe microtubule copolymerized with GTP and Mal3-143 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Microtubule integrity protein mal3, ... | | Authors: | von Loeffelholz, O, Moores, C. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Nucleotide- and Mal3-dependent changes in fission yeast microtubules suggest a structural plasticity view of dynamics.
Nat Commun, 8, 2017
|
|
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
3MS4
 
 | |
3M0Y
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329A in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Takeda, K, Izumori, K, Kamitori, S. | | Deposit date: | 2010-03-03 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the role of Ser329 and the C-terminal region in the catalytic activity of Pseudomonas stutzeri L-rhamnose isomerase
Protein Eng.Des.Sel., 23, 2010
|
|
3M1U
 
 | |
