6WGI
 
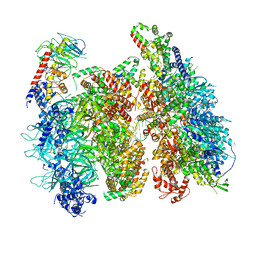 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1C34
 
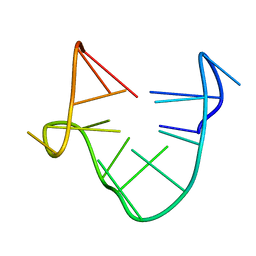 | | SOLUTION STRUCTURE OF A QUADRUPLEX FORMING DNA AND ITS INTERMIDIATE | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Bolton, P.H, Marathias, V.M, Wang, K. | | Deposit date: | 1999-07-24 | | Release date: | 1999-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the potassium-saturated, 2:1, and intermediate, 1:1, forms of a quadruplex DNA.
Nucleic Acids Res., 28, 2000
|
|
2STT
 
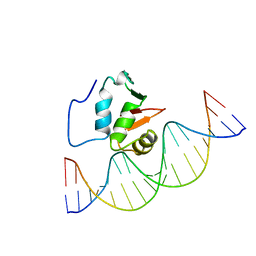 | | SOLUTION NMR STRUCTURE OF THE HUMAN ETS1/DNA COMPLEX, 25 STRUCTURES | | Descriptor: | DNA (5'-D(*TP*CP*GP*AP*AP*CP*TP*TP*CP*CP*GP*GP*CP*TP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*CP*CP*GP*GP*AP*AP*GP*TP*TP*CP*GP*A)-3'), ETS1 | | Authors: | Clore, G.M, Werner, M.H, Gronenborn, A.M. | | Deposit date: | 1996-08-05 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Correction of the NMR structure of the ETS1/DNA complex.
J.Biomol.NMR, 10, 1997
|
|
5GNJ
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*AP*AP*CP*AP*CP*GP*TP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*TP*CP*AP*CP*GP*TP*GP*TP*TP*CP*C)-3'), Transcription factor MYC2 | | Authors: | Lian, T, Xu, Y, Su, X. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Tetrameric Arabidopsis MYC2 Reveals the Mechanism of Enhanced Interaction with DNA.
Cell Rep, 19, 2017
|
|
117D
 
 | |
141D
 
 | |
6ML2
 
 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
3Q5F
 
 | | Crystal structure of the Salmonella transcriptional regulator SlyA in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*CP*TP*TP*AP*GP*CP*AP*AP*GP*CP*TP*AP*AP*TP*TP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*TP*AP*AP*TP*TP*AP*GP*CP*TP*TP*GP*CP*TP*AP*AP*GP*TP*TP*AP*T)-3'), Transcriptional regulator slyA | | Authors: | Dolan, K.T, Duguid, E.M. | | Deposit date: | 2010-12-28 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structures of SlyA Protein, a Master Virulence Regulator of Salmonella, in Free and DNA-bound States.
J.Biol.Chem., 286, 2011
|
|
140D
 
 | | SOLUTION STRUCTURE OF A CONSERVED DNA SEQUENCE FROM THE HIV-1 GENOME: RESTRAINED MOLECULAR DYNAMICS SIMULATION WITH DISTANCE AND TORSION ANGLE RESTRAINTS DERIVED FROM TWO-DIMENSIONAL NMR SPECTRA | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*TP*GP*CP*CP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*GP*GP*CP*AP*AP*GP*CP*T)-3') | | Authors: | Mujeeb, A, Kerwin, S.M, Kenyon, G.L, James, T.L. | | Deposit date: | 1993-09-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra.
Biochemistry, 32, 1993
|
|
197D
 
 | |
4PDI
 
 | |
4GZ2
 
 | | Mus Musculus Tdp2 excluded ssDNA complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*CP*CP*GP*AP*AP*TP*TP*CP*G)-3'), FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Williams, R.S. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of repair of 5'-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1XC8
 
 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-09-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
5IIO
 
 | | Crystal structure of the DNA polymerase lambda binary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
4PDG
 
 | | Crystal structure of a complex between an inhibited LlFpg and a THF containing DNA | | Descriptor: | 2-sulfanyl-1,9-dihydro-6H-purin-6-one, DNA (5'-D(*CP*TP*CP*TP*TP*TP*(3DR)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), ... | | Authors: | Coste, F, Castaing, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zinc finger oxidation of Fpg/Nei DNA glycosylases by 2-thioxanthine: biochemical and X-ray structural characterization.
Nucleic Acids Res., 42, 2014
|
|
6HMS
 
 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
3EPZ
 
 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
1CF7
 
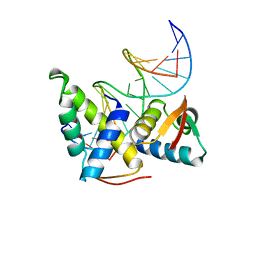 | | STRUCTURAL BASIS OF DNA RECOGNITION BY THE HETERODIMERIC CELL CYCLE TRANSCRIPTION FACTOR E2F-DP | | Descriptor: | DNA (5'-D(*AP*TP*TP*TP*TP*CP*GP*CP*GP*CP*GP*GP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*CP*CP*GP*CP*GP*CP*GP*AP*AP*AP*A)-3'), PROTEIN (TRANSCRIPTION FACTOR DP-2), ... | | Authors: | Zheng, N, Fraenkel, E, Pabo, C.O, Pavletich, N.P. | | Deposit date: | 1999-03-24 | | Release date: | 1999-04-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP.
Genes Dev., 13, 1999
|
|
8HCM
 
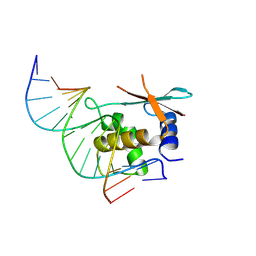 | | zebrafish IRF-11 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*TP*CP*AP*CP*TP*TP*TP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*GP*AP*AP*AP*GP*TP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
8HCL
 
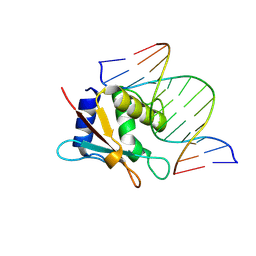 | | zebrafish IRF-10 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*TP*TP*CP*AP*CP*TP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*AP*GP*TP*GP*AP*AP*AP*GP*T)-3'), Interferon regulatory factor 10 | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
3GXQ
 
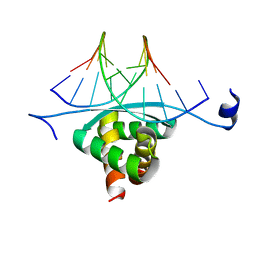 | | Structure of ArtA and DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*AP*TP*GP*AP*CP*AP*TP*G)-3'), DNA (5'-D(*AP*CP*AP*TP*GP*TP*CP*AP*TP*GP*T)-3'), Putative regulator of transfer genes ArtA | | Authors: | Ni, L, Firth, N, Schumacher, M.A. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes.
Nucleic Acids Res., 37, 2009
|
|
1Z9C
 
 | | Crystal structure of OhrR bound to the ohrA promoter: Structure of MarR family protein with operator DNA | | Descriptor: | DNA (29-MER), Organic hydroperoxide resistance transcriptional regulator | | Authors: | Hong, M, Fuangthong, M, Helmann, J.D, Brennan, R.G. | | Deposit date: | 2005-04-01 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure of an OhrR-ohrA Operator Complex Reveals the DNA Binding Mechanism of the MarR Family.
Mol.Cell, 20, 2005
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
1BUA
 
 | |
6BHX
 
 | | B. subtilis SsbA with DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Single-stranded DNA-binding protein A | | Authors: | Dubiel, K.D, Myers, A.R, Satyshur, K.A, Keck, J.L. | | Deposit date: | 2017-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.936 Å) | | Cite: | Structural Mechanisms of Cooperative DNA Binding by Bacterial Single-Stranded DNA-Binding Proteins.
J. Mol. Biol., 431, 2019
|
|
