1B54
 
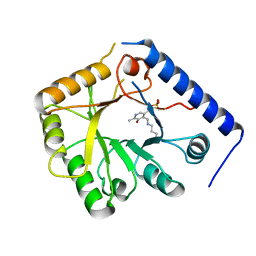 | | CRYSTAL STRUCTURE OF A YEAST HYPOTHETICAL PROTEIN-A STRUCTURE FROM BNL'S HUMAN PROTEOME PROJECT | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, YEAST HYPOTHETICAL PROTEIN | | Authors: | Swaminathan, S, Eswaramoorthy, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-01-12 | | Release date: | 1999-01-27 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a yeast hypothetical protein selected by a structural genomics approach.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1B55
 
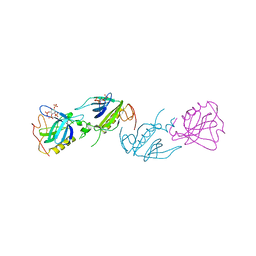 | | PH DOMAIN FROM BRUTON'S TYROSINE KINASE IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, TYROSINE-PROTEIN KINASE BTK, ZINC ION | | Authors: | Djinovic Carugo, K, Baraldi, E, Hyvoenen, M, Lo Surdo, P, Riley, A.M, Potter, B.V.L, O'Brien, R, Ladbury, J.E, Saraste, M. | | Deposit date: | 1999-01-12 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate.
Structure Fold.Des., 7, 1999
|
|
1B56
 
 | | HUMAN RECOMBINANT EPIDERMAL FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Van Tilbeurgh, H, Hohoff, C, Borchers, T, Spener, F. | | Deposit date: | 1999-01-12 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein.
Biochemistry, 38, 1999
|
|
1B57
 
 | | CLASS II FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE IN COMPLEX WITH PHOSPHOGLYCOLOHYDROXAMATE | | Descriptor: | CHLORIDE ION, PHOSPHOGLYCOLOHYDROXAMIC ACID, PROTEIN (FRUCTOSE-BISPHOSPHATE ALDOLASE II), ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 1999-01-12 | | Release date: | 2000-01-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity.
J.Mol.Biol., 287, 1999
|
|
1B58
 
 | |
1B59
 
 | | COMPLEX OF HUMAN METHIONINE AMINOPEPTIDASE-2 COMPLEXED WITH OVALICIN | | Descriptor: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE) | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1B5A
 
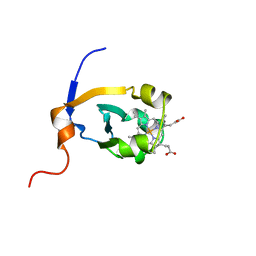 | | RAT FERROCYTOCHROME B5 A CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1B5B
 
 | | RAT FERROCYTOCHROME B5 B CONFORMATION, NMR, 1 STRUCTURE | | Descriptor: | FERROCYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dangi, B, Sarma, S, Yan, C, Banville, D, Guiles, R.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | The origin of differences in the physical properties of the equilibrium forms of cytochrome b5 revealed through high-resolution NMR structures and backbone dynamic analyses.
Biochemistry, 37, 1998
|
|
1B5D
 
 | | DCMP Hydroxymethylase from T4 (Intact) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B5E
 
 | | DCMP HYDROXYMETHYLASE FROM T4 | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, PROTEIN (DEOXYCYTIDYLATE HYDROXYMETHYLASE) | | Authors: | Song, H.K, Sohn, S.H, Suh, S.W. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of deoxycytidylate hydroxymethylase from bacteriophage T4, a component of the deoxyribonucleoside triphosphate-synthesizing complex.
EMBO J., 18, 1999
|
|
1B5F
 
 | | NATIVE CARDOSIN A FROM CYNARA CARDUNCULUS L. | | Descriptor: | PROTEIN (CARDOSIN A), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Frazao, C, Bento, I, Carrondo, M.A. | | Deposit date: | 1999-01-06 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of cardosin A, a glycosylated and Arg-Gly-Asp-containing aspartic proteinase from the flowers of Cynara cardunculus L.
J.Biol.Chem., 274, 1999
|
|
1B5G
 
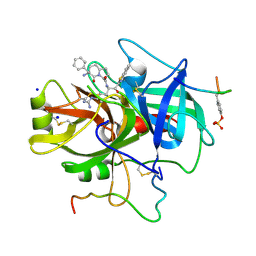 | | HUMAN THROMBIN COMPLEXED WITH NOVEL SYNTHETIC PEPTIDE MIMETIC INHIBITOR AND HIRUGEN | | Descriptor: | ALPHA-THROMBIN, HIRUGEN, SODIUM ION, ... | | Authors: | St Charles, R, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-05-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
1B5H
 
 | |
1B5I
 
 | |
1B5J
 
 | |
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1B5L
 
 | | OVINE INTERFERON TAU | | Descriptor: | INTERFERON TAU, SULFATE ION | | Authors: | Radhakrishnan, R, Walter, L.J, Subramaniam, P.S, Johnson, H.J, Walter, M.R. | | Deposit date: | 1999-01-07 | | Release date: | 1999-05-18 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ovine interferon-tau at 2.1 A resolution.
J.Mol.Biol., 286, 1999
|
|
1B5M
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rivera, M, White, S.P, Zhang, X. | | Deposit date: | 1996-11-07 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c.
Biochemistry, 35, 1996
|
|
1B5N
 
 | | NMR STRUCTURE OF PSP1, PLASMATOCYTE-SPREADING PEPTIDE FROM PSEUDOPLUSIA INCLUDENS | | Descriptor: | PROTEIN (PLASMATOCYTE-SPREADING PEPTIDE) | | Authors: | Volkman, B.F, Clark, K.D, Anderson, M.E, Pech, L.L, Markley, J.L, Strand, M.R. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the insect cytokine peptide plasmatocyte-spreading peptide 1 from Pseudoplusia includens.
J.Biol.Chem., 274, 1999
|
|
1B5O
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE SINGLE MUTANT 1 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-07 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1B5P
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-07 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
1B5Q
 
 | | A 30 ANGSTROM U-SHAPED CATALYTIC TUNNEL IN THE CRYSTAL STRUCTURE OF POLYAMINE OXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, ... | | Authors: | Binda, C, Coda, A, Angelini, R, Federico, R, Ascenzi, P, Mattevi, A. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase.
Structure Fold.Des., 7, 1999
|
|
1B5S
 
 | | DIHYDROLIPOYL TRANSACETYLASE (E.C.2.3.1.12) CATALYTIC DOMAIN (RESIDUES 184-425) FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Izard, T, Aevarsson, A, Allen, M.D, Westphal, A.H, Perham, R.N, De Kok, A, Hol, W.G. | | Deposit date: | 1999-01-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1B5T
 
 | | ESCHERICHIA COLI METHYLENETETRAHYDROFOLATE REDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MERCURY (II) ION, PROTEIN (METHYLENETETRAHYDROFOLATE REDUCTASE) | | Authors: | Guenther, B.D, Sheppard, C.A, Tran, P, Rozen, R, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia.
Nat.Struct.Biol., 6, 1999
|
|
1B5U
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANT | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
