6KTZ
 
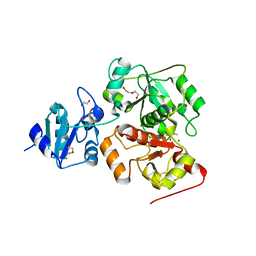 | | The complex structure of EanB/C412S with hercynine | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
6KTV
 
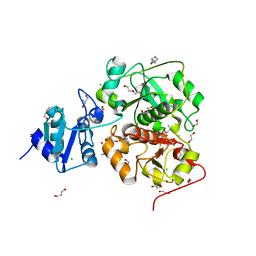 | | The structure of EanB complex with hercynine and persulfided Cys412 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, CHLORIDE ION, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
1ZB7
 
 | | Crystal Structure of Botulinum Neurotoxin Type G Light Chain | | Descriptor: | CITRATE ANION, ZINC ION, neurotoxin | | Authors: | Arndt, J.W, Yu, W, Bi, F, Stevens, R.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of botulinum neurotoxin type g light chain: serotype divergence in substrate recognition
Biochemistry, 44, 2005
|
|
5UQ1
 
 | |
5VR0
 
 | |
6HVF
 
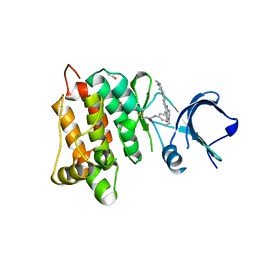 | | Kinase domain of cSrc in complex with compound 29B | | Descriptor: | 1,2-ETHANEDIOL, Proto-oncogene tyrosine-protein kinase Src, ~{N}-[3-[3-ethyl-6-[4-(4-methylpiperazin-1-yl)phenyl]-4-oxidanylidene-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]phenyl]prop-2-enamide | | Authors: | Keul, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of osimertinib-resistant epidermal growth factor receptor EGFR-T790M/C797S.
Chem Sci, 10, 2019
|
|
6DTD
 
 | |
6HZB
 
 | |
6HZC
 
 | |
5VKC
 
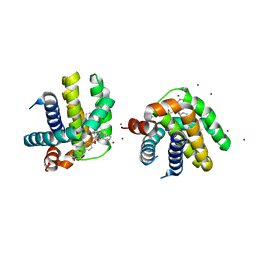 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
6I6F
 
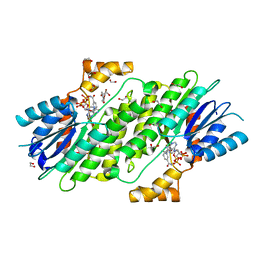 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6I6V
 
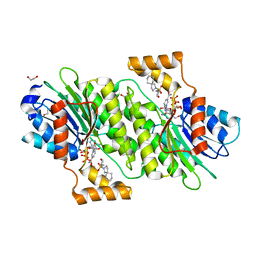 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(3~{R})-oxan-3-yl]methylsulfonyl]-2-azaspiro[4.5]decane, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5W8C
 
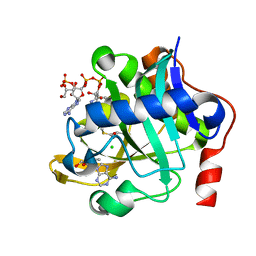 | | The structure of a COA-dependent acyl-homoserine lactone synthase, BjaI, with MTA and isovaleryl-CoA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Autoinducer synthase, CHLORIDE ION, ... | | Authors: | Dong, S.-H, Nair, S.K. | | Deposit date: | 2017-06-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the substrate specificity of quorum signal synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W8D
 
 | |
5W86
 
 | |
5WFS
 
 | |
5WIH
 
 | |
5WNS
 
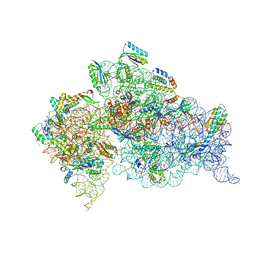 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S Ribosomal RNA rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | DeMirci, H. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2'-O-methylation in mRNA disrupts tRNA decoding during translation elongation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5W85
 
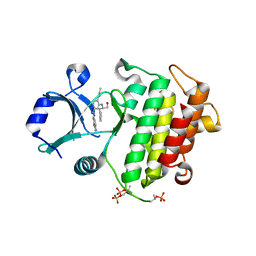 | | CRYSTAL STRUCTURE OF IRAK-4 WITH A 4,6-DIAMINONICOTINAMIDE INHIBITOR (COMPOUND NUMBER 9) | | Descriptor: | 6-[(1,3-benzothiazol-6-yl)amino]-4-{[(2S)-1-hydroxy-3-phenylpropan-2-yl]amino}-N-methylpyridine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Sack, J.S. | | Deposit date: | 2017-06-21 | | Release date: | 2017-10-11 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and structure-based design of 4,6-diaminonicotinamides as potent and selective IRAK4 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5WF0
 
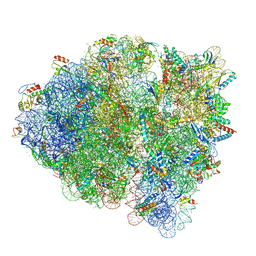 | |
5WFK
 
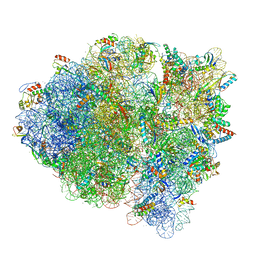 | |
5WJY
 
 | |
5WJU
 
 | | Cryo-EM structure of B. subtilis flagellar filaments A39V, N133H | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
1YSO
 
 | | YEAST CU, ZN SUPEROXIDE DISMUTASE WITH THE REDUCED BRIDGE BROKEN | | Descriptor: | COPPER (I) ION, YEAST CU, ZN SUPEROXIDE DISMUTASE, ... | | Authors: | Parge, H.E, Crane, B.R, Tsang, J, Tainer, J.A. | | Deposit date: | 1995-12-21 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual trigonal-planar copper configuration revealed in the atomic structure of yeast copper-zinc superoxide dismutase.
Biochemistry, 35, 1996
|
|
1YLI
 
 | |
