8XDU
 
 | |
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8YQP
 
 | | Crystal structure of HylD1 in complex with MEP | | Descriptor: | 2-ethoxycarbonylbenzoic acid, Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
8XW3
 
 | |
8XDS
 
 | |
9AZB
 
 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
8Z1L
 
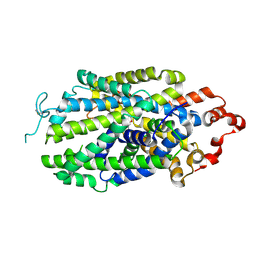 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 2024
|
|
9EUR
 
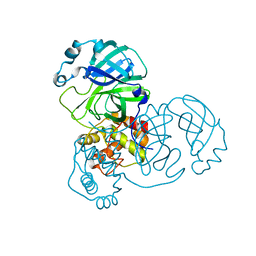 | | Mpro WT from SARS-CoV-2 with 298Q mutation | | Descriptor: | Replicase polyprotein 1a | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8YKU
 
 | |
8YUD
 
 | |
8XDY
 
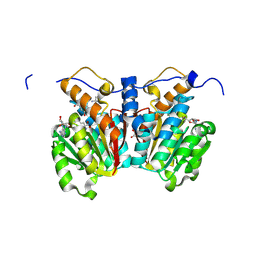 | | O-methyltransferase from Lycoris longituba M52A variant complexed with Mg, SAH, and 3,4-dihydroxybenzaldehyde | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Saw, Y.Y.H, Nakashima, Y, Morita, H. | | Deposit date: | 2023-12-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Catalytic Mechanism of Amaryllidaceae O-Methyltransferases
Acs Catalysis, 2024
|
|
8XWK
 
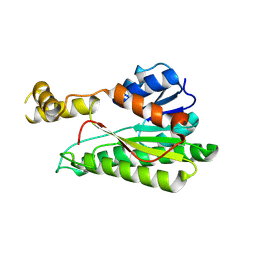 | |
8YL9
 
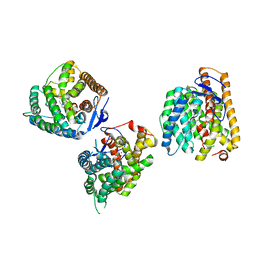 | | Crystal structures of terpene synthases complexed with a substrate mimic | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE, ... | | Authors: | Xu, M, Ma, M. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights Into the Terpene Cyclization Domains of Two Fungal Sesterterpene Synthases and Enzymatic Engineering for Sesterterpene Diversification.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
9EPL
 
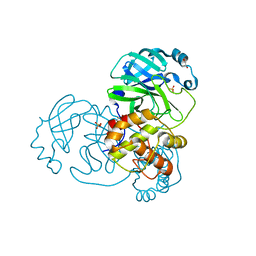 | | Mpro from SARS-CoV-2 with 298Q mutation | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Non-structural protein 11, ... | | Authors: | Plewka, J, Lis, K, Czarna, A, Pyrc, K, Kantyka, T, Chykunova, Y. | | Deposit date: | 2024-03-18 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8Y6Q
 
 | | Structure of the Dark/Dronc complex | | Descriptor: | Apaf-1 related killer DARK, Caspase Dronc | | Authors: | Tian, L, Li, Y, Shi, Y. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Dark and Dronc activation in Drosophila melanogaster.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9ER3
 
 | |
9ATJ
 
 | | Crystal structure of MERS 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9EWM
 
 | | Mpro from SARS-CoV-2 with R4Q R298Q double mutations | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
9EWO
 
 | | Mpro from SARS-CoV-2 with R4A R298A double mutations | | Descriptor: | Non-structural protein 11, SULFATE ION | | Authors: | Plewka, J, Lis, K, Chykunova, Y, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
8YOP
 
 | | Cryo-EM structure of the human 80S ribosome with 4 um Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Denk, T, Cheng, J. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XDV
 
 | |
9B0Z
 
 | |
8XCI
 
 | |
9EWN
 
 | | Mpro from SARS-CoV-2 with 4Q mutation | | Descriptor: | Non-structural protein 11 | | Authors: | Plewka, J, Lis, K, Czarna, A, Kantyka, T, Pyrc, K. | | Deposit date: | 2024-04-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | SARS-CoV-2 M pro oligomerization as a potential target for therapy.
Int.J.Biol.Macromol., 267, 2024
|
|
